Abstract
The most common recurrent cytogenetic abnormalities in T-lymphoblastic leukemia (T-acute lymphoblastic leukemia [T-ALL]) involve T-cell receptor (TCR) loci and a variety of partner genes, including HOX11, HOX11L2, MYC, and TAL1. In this report, we present a rare case involving simultaneous translocation of the TCR α/δ loci with different partner loci (Xq22 and 12p13); this resulted in a poor prognosis. Chromosomal analysis showed 46,Y,t(X;14)(q22;q11.2),t(12;14)(p13;q11.2) and FISH analysis by using a T-cell receptor alpha delta DNA probe, Split Signal (DakoCytomation, Denmark), showed translocations at the same TCR α/δ locus on both chromosomes. FISH with 2 bacterial artificial chromosome clones showed break apart signal, which suggests involvement of the IRS4 gene. To our knowledge, this is the first report of T-ALL in which both TCR α/δ loci were translocated with different partner loci, and 1 of the partner loci, Xq22, was a rare translocation partner locus that included IRS4 gene.
T-lymphoblastic leukemia (T-cell acute lymphoblastic leukemia [T-ALL]), a highly malignant cancer derived from T-cell progenitors, comprises 15% of acute lymphoblastic leukemia cases [1, 2]. T-ALL is a genetically heterogeneous disease with translocations that usually involve recombination between T-cell receptor (TCR) loci and several different partner genes [3]. Most of these translocations result in deregulation of the partner genes that are located near TCR regulatory elements [4]. The most common partner genes are HOX11, HOX11L2, MYC, and TAL1. Consequently, these partner genes, rather than the TCR, may play important roles in T-cell leukemogenesis. Translocations involving TCR loci are found in about 35% of T-ALL cases, and unidentified partner genes are involved in 5-10% of cases [5]. This report presents a case of T-ALL in which the leukemic cells showed the simultaneous chromosomal abnormalities t(X;14)(q22;q11.2) and t(12;14)(p13;q11.2). To our knowledge, this simultaneous translocation has not been previously reported, and Xq22 has been reported as a partner locus only once in childhood T-ALL [3].
A 27-yr-old man was admitted to our hospital complaining of headache, dizziness, nausea, and vomiting, which began 3 days before admission. No palpable lymph-node enlargement or hepatosplenomegaly was detected on physical examination. Laboratory findings were as follows: white blood cell (WBC) count, 49.5×109/L; hemoglobin level, 161 g/L; and platelet count, 36×109/L. A peripheral blood smear showed that 95% of WBCs were blasts (Fig. 1A). The serum lactate dehydrogenase (LDH) level was markedly elevated to 13,022 IU/L. The patient's bone marrow was nearly packed with small-to-medium sized leukemic cells with a high nuclear/cytoplasmic ratio, which was calculated as 93.2% (Fig. 1B, C). A biopsy showed hypercellularity with diffuse infiltration of immature blast cells (Fig. 1D). Flow cytometry revealed that the leukemic blasts with intermediate CD45 expression and low side scatter (SSC) were positive for CD2, CD7, CD5, and CD3, and negative for TdT, CD34, CD13, CD33, CD10, CD19, CD20, and CD22. Therefore, the patient was diagnosed with T-ALL.
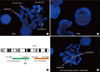
The patient's karyotype was 46,Y,t(X;14)(q22;q11.2),t(12;14)(p13;q11.2) (Fig. 2). FISH analysis was performed to determine the breakpoints using a T-cell receptor alpha delta (TCRAD) DNA Probe, Split Signal (DakoCytomation, Glostrup, Denmark), and it revealed 2 red and 2 green abnormal signal patterns in 89% of the cells, suggesting TCR α/δ rearrangements in both chromosomes. To map the Xq22 breakpoint, 2 bacterial artificial chromosome (BAC) clones, RP11-815E21 and RP11-105F23 (Empire Genomics, Buffalo, NY, USA), were used [3], and break-apart signal patterns were obtained, suggesting insulin receptor substrate (IRS)4 gene translocation (Fig. 3). Additional FISH analyses by using the LSI BCR/ABL Dual Color, Dual Fusion Translocation Probe (Vysis/Abbott Molecular, Des Plaines, IL, USA), the TEL/AML1 ES Dual Color Translocation Probe (Vysis/Abbott Molecular), the LSI p16(9p21)/CEP 9 Dual Color Probe (Vysis/Abbott Molecular), the LSI MLL Dual Color, Break Apart Rearrangement Probe (Vysis/Abbott Molecular), and the LSI MYC Dual Color, Break Apart Rearrangement Probe (Vysis/Abbott Molecular) showed no abnormal signals.
The patient was treated with hyper-CVAD (cyclophosphamide, vincristine, adriamycin, and dexamethasone) as the first course of induction chemotherapy. Complete remission was achieved after 1 month and was confirmed by normal TCRAD FISH signals. At that time, the patient's karyotype was 46,XY [20]. Next, the patient was treated with high-dose methotrexate/cytarabine (HD MTx/Ara-C) as the first course of consolidation therapy, hyper-CVAD as the second consolidation therapy, and then allogeneic peripheral blood stem cell transplantation (allo-PBSCT) was performed. However, a central nervous system (CNS) relapse was diagnosed 1 yr later, and the patient died of sepsis while receiving induction chemotherapy 1 month after relapse.
T-ALL is a malignant proliferation of T-lymphoid blasts, and 50-70% of cases have abnormal karyotypes [2, 6]. The chromosomal rearrangements in T-ALL typically involve breakpoints in bands where TCR genes are located, including 14q11 (TCR-α and TCR-δ), 7q32-36 (TCR-β), and 7p15 (TCR-γ) [2, 7]. During V(D)J recombination in T-cell development, several genes transcribed at an early stage of thymocyte development are in an "open" chromatin configuration and are vulnerable to the action of recombinases. Consequently, illegitimate recombination may place target partner genes next to TCR regulatory sequences. This may lead to their aberrant expression in developing thymocytes, thus leading to T-ALL, with differentiation blocked at various stages of maturation [5]. Commonly involved partner genes of TCR loci include the transcription factors HOX11 (TLX1) (10q24), HOX11L2 (TLX3) (5q35), MYC (8q24.1), TAL1 (1p32), TAL2 (9q32), LYL1 (19p13), LMO1 (11p15), and LMO2 (11p13), and the cytoplasmic tyrosine kinase LCK (1p34.3-35) [6, 8-10].
Simultaneous rearrangements targeting TCR-β and -α loci are observed in 4% of patients with T-ALL, possibly reflecting a higher susceptibility to errors in V(D)J recombination [11]. In this study, we described a case of T-ALL with simultaneous translocation of both TCR α/δ loci (14q11) to different partner loci (Xq22 and 12p13). From a molecular perspective, Xq22 and 12p13 were juxtaposed with the TCR α/δ loci (14q11). Breakpoints within the TCR α/δ locus occur in more than 20% of T-ALL cases with abnormal karyotypes, and the mechanism underlying these translocations most likely involves illegitimate V(D)J recombination [4, 12, 13].
In several T-ALL cases, t(12;14)(p13;q11) has been observed in association with the CCND2 gene, which is thought to stimulate cellular proliferation via cell cycle activation [1, 4]. The Xq22 locus is a rare partner gene, and to our knowledge, has been reported only once by Karrman et al. [3] in a pediatric case of T-ALL in which Xq22 was translocated to 7q34 (TCR-β). There is no report of adult T-ALL involving the translocation of Xq22 and TCR α/δ. The molecular genetic characterization of t(X:7) revealed the IRS4 gene [3, 14].
Although an additional study was not available in our case, FISH using the RP11-815E21 and RP11-105F23 BAC probes (Empire Genomics) at Xq22.3 showed a break-apart signal pattern, suggesting IRS4 gene translocation. The exact role of IRS4 remains unclear; however, IRS proteins mediate signaling from insulin and insulin-like growth factor 1 receptors, and have important effects on cell growth and survival [15].
In rarely observed cytogenetic abnormalities, it is difficult to uncover any relationship with prognosis. In a UK-US collaborative group study of 356 adult T-ALL cases, the remission rate was over 90%, and the 5-yr survival rate was approximately 50% [16]. However, considering that in the case reported by Karrman et al. [3] the patient relapsed and died about 2 yr after diagnosis, and the outcome in our case, where the patient relapsed and died about 1 yr after the initial diagnosis, IRS4 gene involvement may be related to a poor prognosis.
In conclusion, this is the first case report of T-ALL in which both TCR α/δ loci (14q11) were translocated simultaneously to different partner loci. One of these partner loci, Xq22, was rare and was found to involve IRS4, which may have an important effect on cell growth and survival, and to be associated with a poor prognosis.
Figures and Tables
Fig. 1
T-lymphoblastic leukemia. Peripheral blood smear containing leukemic cells (Wright's stain, ×200) (A). Bone marrow aspirate smear showing leukemic cells that are small-to-medium sized, with a high nuclear-to-cytoplasmic ratio (Wright-Giemsa stain, ×200 (B) and ×1,000 (C)). Biopsy section showing hypercellularity with heavy infiltration of immature cells (H&E, ×100) (D).
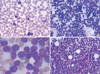
Fig. 2
Karyotype at diagnosis showing 46,Y,t(X;14)(q22;q11.2),t(12;14)(p13;q11.2). Arrows indicate the rearranged chromosomes.

Fig. 3
FISH at initial diagnosis using the TCRAD DNA Probe, Split Signal (DakoCytomation, Glostrup, Denmark), which showed 2 red and 2 green abnormal signals in metaphase (A) and interphase (B). A schematic diagram of the positions of RP11-815E21 and RP11-105F23 at Xq22.3 (C). Start and end positions were acquired from NCBI36/hg18 assembly. FISH using RP11-815E21 and RP11-105F23 showing 1 red and 1 green break-apart signal (D).
Abbreviation: TCRAD, T-cell receptor alpha delta.
References
1. Clappier E, Cuccuini W, Cayuela JM, Vecchione D, Baruchel A, Dombret H, et al. Cyclin D2 dysregulation by chromosomal translocations to TCR loci in T-cell acute lymphoblastic leukemias. Leukemia. 2006. 20:82–86.

2. Douet-Guilbert N, Morel F, Le Bris MJ, Herry A, Le Calvez G, Marion V, et al. Cytogenetic studies in T-cell acute lymphoblastic leukemia (1981-2002). Leuk Lymphoma. 2004. 45:287–290.

3. Karrman K, Kjeldsen E, Lassen C, Isaksson M, Davidsson J, Andersson A, et al. The t(X;7)(q22;q34) in paediatric T-cell acute lymphoblastic leukaemia results in overexpression of the insulin receptor substrate 4 gene through illegitimate recombination with the T-cell receptor beta locus. Br J Haematol. 2009. 144:546–551.

4. Karrman K, Andersson A, Björgvinsdóttir H, Strömbeck B, Lassen C, Olofsson T, et al. Deregulation of cyclin D2 by juxtaposition with T-cell receptor alpha/delta locus in t(12;14)(p13;q11)-positive childhood T-cell acute lymphoblastic leukemia. Eur J Haematol. 2006. 77:27–34.

5. Graux C, Cools J, Michaux L, Vandenberghe P, Hagemeijer A. Cytogenetics and molecular genetics of T-cell acute lymphoblastic leukemia: from thymocyte to lymphoblast. Leukemia. 2006. 20:1496–1510.

6. Borowitz M, Chan J. Swerdlow SH, Campo E, editors. T lymphoblastic Leukeaemia/lymphoma. WHO classification of tumours of haematopoietic and lymphoid tissues. 2008. 4th ed. Lyon: IARC Press;176–178.
7. Gesk S, Martín-Subero JI, Harder L, Luhmann B, Schlegelberger B, Calasanz MJ, et al. Molecular cytogenetic detection of chromosomal breakpoints in T-cell receptor gene loci. Leukemia. 2003. 17:738–745.

9. Karrman K, Forestier E, Heyman M, Andersen MK, Autio K, Blennow E, et al. Clinical and cytogenetic features of a population-based consecutive series of 285 pediatric T-cell acute lymphoblastic leukemias: rare T-cell receptor gene rearrangements are associated with poor outcome. Genes Chromosomes Cancer. 2009. 48:795–805.

10. Van Vlierberghe P, Pieters R, Beverloo HB, Meijerink JP. Molecular-genetic insights in paediatric T-cell acute lymphoblastic leukaemia. Br J Haematol. 2008. 143:153–168.

11. Raimondi SC, editor. T-lineage acute lymphoblastic leukemia (T-ALL). Atlas Genet Cytogenet Oncol Haematol. Updated on May 2007. http://atlasgeneticsoncology.org/Anomalies/TALLID1374.html.

12. Inaba T, Murakami S, Oku N, Itoh K, Ura Y, Nakanishi S, et al. Translocation between chromosomes 8q24 and 14q11 in T-cell acute lymphoblastic leukemia. Cancer Genet Cytogenet. 1990. 49:69–74.

13. Mandac I, Kolonić SO, Vrhovac R, Lasan-Trcić R, Jakelić-Pitesa J, Kardum-Skelin I. T-lymphoblastic lymphoma with an unusual t(8;14)(q24;q11)-case report. Coll Antropol. 2010. 34:265–269.
14. Fantin VR, Keller SR, Lienhard GE, Wang LM. Insulin receptor substrate 4 supports insulin- and interleukin 4-stimulated proliferation of hematopoietic cells. Biochem Biophys Res Commun. 1999. 260:718–723.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download