Abstract
Background
The identification of molds in clinical laboratories is largely on the basis of phenotypic criteria, the classification of which can be subjective. Recently, molecular methods have been introduced for identification of pathogenic molds in clinical settings. Here, we employed comparative sequence analysis to identify molds.
Methods
A total of 47 clinical mold isolates were used in this study, including Aspergillus and Trichophyton. All isolates were identified by phenotypic properties, such as growth rate, colony morphology, and reproductive structures. PCR and direct sequencing, targeting the internal transcribed spacer (ITS) region, the D1/D2 region of the 28S subunit, and the β-tubulin gene, were performed using primers described previously. Comparative sequence analysis by using the GenBank database was performed with the basic local alignment search tool (BLAST) algorithm.
Results
For Aspergillus, 56% and 67% of the isolates were identified to the species level by using ITS and β-tubulin analysis, respectively. Only D1/D2 analysis was useful for Trichophyton identification, with 100% of isolates being identified to the species level. Performances of ITS and D1/D2 analyses were comparable for species-level identification of molds other than Aspergillus and Trichophyton. In contrast, the efficacy of β-tubulin analysis was limited to genus identification because of the paucity of database information for this gene.
The epidemiology and etiology of invasive fungal infections have changed over recent decades. Mold infections are more frequently encountered in association with increasing numbers of immunocompromised patients, and molds other than Aspergillus fumigatus, including species not previously recognized as pathogens, have emerged [1, 2]. The appearance of organisms, such as the Fusarium species and the Zygomycetes, with variable susceptibilities to conventional antifungal agents underscores the importance of correct identification [3].
Clinical laboratories are often challenged with mold identification. In contrast with bacterial or Candida species, which are identified on the basis of biochemical properties, mold identification is largely based on phenotypic criteria. Related species or phenotypic variants may be misidentified and rare species may remain unidentified. As a result, molecular methods have been developed to overcome these problems, and comparative sequence analysis is now considered the gold standard identification technique [4, 5].
The internal transcribed spacer (ITS) region is the most commonly used target for sequencing in clinical laboratories because of the following benefits: (i) multiple copies of the ribosomal gene are present in all organisms, enabling sensitive detection by PCR, and (ii) the ITS region contains both highly conserved and variable regions, and is therefore, the optimal target for developing specific PCR primers that discriminate among closely related species [6-9]. However, the ITS region might not provide species-level resolution for all species. In such cases, other targets such as the D1/D2 region of the 28S subunit, the β-tubulin gene, or the translation elongation factor gene may prove useful, depending on the genus in question. In this study, we used molecular methods for mold identification and compared the performances of the ITS region, the D1/D2 region, and the β-tubulin gene as amplification targets for comparative sequence analysis.
Forty-seven preserved isolates, which were previously obtained from clinical specimens, were cultivated to evaluate the usefulness of each locus for mold identification. Isolate selection criteria were as follows: (i) isolates most frequently recovered in our laboratory, (ii) medically important isolates recovered infrequently, or (iii) isolates unidentifiable by phenotypic methods. The genera included in this study were Absidia (1), Acremonium (1), Aspergillus (9), Cladosporium (2), Cunninghamella (1), Exophiala (1), Fusarium (2), Paecilomyces (3), Microsporum (1), Penicillium (3), Rhizomucor (1), Rhizopus (1), Scopulariopsis (1), Scedosporium (1), Sporothrix (2), Trichophyton (9), and unidentifiable molds (8).
Clinical specimens were cultured on universal media (Sabouraud dextrose agar) and/or selective media (Mycogel agar) for a maximum of 3 weeks, according to the type of specimen. Specimens were incubated at 30℃ for the first 2 days and then at 25℃. Isolates were sub-cultured for identification on Sabouraud dextrose agar or potato dextrose agar. Presumptive thermally dimorphic fungi were grown on Sabouraud dextrose agar at 25℃ and brain heart infusion agar at 37℃. Conventional identification was made according to macro- and micro-morphologic criteria.
DNA was extracted using the MagNA Pure LC DNA Isolation Kit III (bacteria, fungi) (Roche Diagnostics, Mannheim, Germany). Fungal mycelium with a surface area of 2-4 cm2 was obtained, added to phosphate buffered saline, and samples were prepared according to the manufacturer's recommendations. Briefly, approximately 100 µL of sample was mixed with 130 µL of bacterial lysis buffer and 20 µL of proteinase K, incubated at 95℃ for 10 min and then cooled. After sample preparation DNA was isolated using magnetic-bead technology, according to the manufacturer's instructions.
PCR was performed using a thermal cycler (Model 9700; Applied Biosystems, Foster City, CA, USA), and amplified products were sequenced using the BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems) and an ABI Prism 3100 Genetic Analyzer (Applied Biosystems). Primer sequences used in this study were the same as those given in the Clinical and Laboratory Standard Institute (CLSI) guidelines: (i) sequences of the ITS region covering ITS1, 5.8S, and ITS2 were amplified using ITS-1/ITS-4 and ITS-5/ITS-4 primer sets; (ii) sequences of the D1/D2 region were amplified using NL-1/NL-4 primers; and (iii) β-tubulin gene sequences were amplified using Bt2a/T1 primers.
The amplified sequences were compared with the GenBank (NCBI) database using the basic local alignment search tool (BLAST) algorithm. Using a calculated percent identity score, specimens were assigned to a genus and species, as previously described [6, 7]. Briefly, a sequence was assigned to a species if the best matching reference sequence showed ≥98% homology and the next best matching reference species showed 0.8%, or less, sequence homology. A sequence was assigned to the genus level on the basis of 95% to 98% homology to the best matching sequence or ≥98% homology with sequence entries for several species from the same genus. 'No identification' was defined as <95% homology with the best matching reference sequence or a sequence homology >95% with various genera. Discrepant results between phenotypic and sequence-based identification methods were resolved by repeating the sequencing and re-evaluation of the phenotypic method.
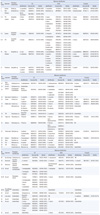
Phenotypic identification and sequencing results for the 3 targets are shown in Table 1. For Aspergillus, 56% (5/9 isolates) and 67% (6/9 isolates) were identified to the species level by ITS and β-tubulin analysis, respectively. None of the isolates was identified to the species level by D1/D2 analysis alone. Only D1/D2 analysis enabled species-level identification for Trichophyton isolates, with 100% of isolates being identified to the species level. ITS and D1/D2 targets yielded comparable performances in identification of molds, other than Aspergillus and Trichophyton.
There was a discrepancy between phenotypic identification and ITS analysis at the genus level for some isolates. Isolate 14 was identified as a Cunninghamella bertholletiae based on typical microscopic features, such as non-septate hyphae, sporangiophores, terminal vesicles, sporangioles, and the knowledge that C. bertholletiae is the only known human pathogen. However, ITS analysis matched the isolate to Nectria mauritiicola. D1/D2 analysis assigned the isolate to the Cunninghamella genus, although species-level identification was not possible using this target. Isolate 19, phenotypically identified as a Paecilomyces species, was re-classified into Geosmithia argillacea on the basis of our molecular analyses. Interestingly, isolate 44, phenotypically unidentifiable, was also classified as G. argillacea. The 2 isolates were recovered from a trans-tracheal aspirate and lung tissue, respectively. Isolate 30 was phenotypically identified as Sporothrix schenkii, while D1/D2 analysis matched the isolate to Saccharomycopsis fibuligera. However, after re-evaluating the phenotypic method, the isolate was finally confirmed as S. schenkii exhibiting thermal dimorphism.
Five of 8 phenotypically unidentifiable isolates were newly identified as Coniosporium sp. (isolate 43), G. argillacea (isolate 44), Phialophora sp. (isolate 45), and Trichophyton rubrum (isolates 46, 47) by sequence analysis. The 3 remaining isolates were still unidentifiable by sequence analysis.
In this study, we employed a molecular method for mold identification and compared the performance of the 3 commonly used amplification targets. Using this method, 2 genera previously unidentified in a clinical laboratory were discovered, Coniosporium and Geosmithia. The Coniosporium species isolate was a slow-growing, black pigmented fungus recovered from a toenail, and showed arthroconidia microscopically. Coniosporium, which is known to colonize plants, has been reported in the literature as a human pathogen recovered from a superficial skin lesion [10]. The isolates identified as G. argillacea grew as whitish to olive colonies and had phialides, which were difficult to distinguish from those of Penicillium or Paecilomyces. Geosmithia is a polyphyletic genus created to accommodate Penicillium species that do not produce green colonies. According to recent reports [11, 12], G. argillacea can colonize the respiratory tract of cystic fibrosis patients, although it was not found to be associated with exacerbation of the disease. One of our isolates was obtained from the trans-tracheal aspirate of an acute lymphoblastic leukemia patient with influenza H1N1 infection, and the other isolate was obtained from the lung tissue of a patient with chronic cavitary pulmonary aspergillosis. Additional research is required to determine the clinical implications of colonization with G. argillacea.
ITS and D1/D2 region analyses performed well for identification of most isolates in this study. However, D1/D2 analysis was not appropriate for species-level identification of Aspergillus species, and this finding is consistent with the results of a previous study [13]. In contrast to D1/D2 analysis, where all species yielded 100% identical sequence data for at least one molecular sibling (closely related but different taxa), the ITS analysis distinguished some of the species (A. fumigates, A. terreus) from their molecular siblings. The β-tubulin gene was also helpful for some species (A. fumigatus, A. terreus, A. sydowii). In contrast, D1/D2 analysis was more appropriate than that of ITS for identifying Trichophyton species. Interestingly, isolates morphologically identified as T. mentagrophytes were re-classified into T. interdigitale (Arthroderma vanbreuseghemii) after analysis of the D1/D2 region, according to the current taxonomy suggested by Graser et al. [14]. The sequence of these isolates was identical to the neotype of T. interdigitale, CBS 428.63 (AF506033), but not with the neotype of T. mentagrophytes, CBS 318.56 (AY185126). Although the naming of the T. mentagrophytes complex has been a topic of debate for years, use of T. mentagrophytes rather than T. interdigitale could result in confusion, an issue recently raised in the literature [15, 16]. A consensus on the taxonomy of the T. mentagrophytes complex must be reached as soon as possible.
ITS and D1/D2 analyses yielded comparable performances for identification of species other than Aspergillus and Trichophyton. β-tubulin analysis was limited to genus-level identification due of the paucity of database information available for this gene. Since there is a variety of reference sequences deposited in the public database, the ITS region may be the most appropriate primary sequencing target, except in the case of Aspergillus, as recommended by the CLSI [17]. Analysis of the D1/D2 region or β-tubulin gene could be used for further resolution, and the decision to use additional targets should be based on clinical implications and laboratory policies, since the relevance of species-level identification has only been determined for a limited number of genera [18, 19]. In summary, molecular methods are useful for mold identification, although the identification performance of each locus varied according to genus. Thus, a tailored approach is recommended when selecting amplification targets for molecular identification of molds.
Figures and Tables
References
2. Malani AN, Kauffman CA. Changing epidemiology of rare mould infections: implications for therapy. Drugs. 2007. 67:1803–1812.
3. Nucci M. Emerging moulds: Fusarium, Scedosporium and Zygomycetes in transplant recipients. Curr Opin Infect Dis. 2003. 16:607–612.

4. Summerbell RC, Levesque CA, Seifert KA, Bovers M, Fell JW, Diaz MR, et al. Microcoding: the second step in DNA barcoding. Philos Trans R Soc Lond B Biol Sci. 2005. 360:1897–1903.

5. Balajee SA, Borman AM, Brandt ME, Cano J, Cuenca-Estrella M, Dannaoui E, et al. Sequence-based identification of Aspergillus, fusarium, and mucorales species in the clinical mycology laboratory: where are we and where should we go from here? J Clin Microbiol. 2009. 47:877–884.
6. Ciardo DE, Schar G, Altwegg M, Bottger EC, Bosshard PP. Identification of moulds in the diagnostic laboratory--an algorithm implementing molecular and phenotypic methods. Diagn Microbiol Infect Dis. 2007. 59:49–60.

7. Ciardo DE, Lucke K, Imhof A, Bloemberg GV, Böttger EC. Systematic internal transcribed spacer sequence analysis for identification of clinical mold isolates in diagnostic mycology: a 5-year study. J Clin Microbiol. 2010. 48:2809–2813.

8. Iwen PC, Hinrichs SH, Rupp ME. Utilization of the internal transcribed spacer regions as molecular targets to detect and identify human fungal pathogens. Med Mycol. 2002. 40:87–109.

9. Atkins SD, Clark IM. Fungal molecular diagnostics: a mini review. J Appl Genet. 2004. 45:3–15.
10. Li DM, de Hoog GS, Saunte DM, van den Ende AH, Chen XR. Coniosporium epidermidis sp. nov., a new species from human skin. Stud Mycol. 2008. 61:131–136.

11. Barton RC, Borman AM, Johnson EM, Houbraken J, Hobson RP, Denton M, et al. Isolation of the fungus Geosmithia argillacea in sputum of people with cystic fibrosis. J Clin Microbiol. 2010. 48:2615–2617.
12. Giraud S, Pihet M, Razafimandimby B, Carrere J, Degand N, Mely L, et al. Geosmithia argillacea: an emerging pathogen in patients with cystic fibrosis. J Clin Microbiol. 2010. 48:2381–2386.
13. Hinrikson HP, Hurst SF, Lott TJ, Warnock DW, Morrison CJ. Assessment of ribosomal large-subunit D1-D2, internal transcribed spacer 1, and internal transcribed spacer 2 regions as targets for molecular identification of medically important Aspergillus species. J Clin Microbiol. 2005. 43:2092–2103.
14. Graser Y, Kuijpers AF, Presber W, De Hoog GS. Molecular taxonomy of Trichophyton mentagrophytes and T. tonsurans. Med Mycol. 1999. 37:315–330.

15. Sun PL, Hsieh HM, Ju YM, Jee SH. Molecular characterization of dermatophytes of the Trichophyton mentagrophytes complex found in Taiwan with emphasis on their correlation with clinical observations. Br J Dermatol. 2010. 163:1312–1318.

16. Heidemann S, Monod M, Graser Y. Signature polymorphisms in the internal transcribed spacer region relevant for the differentiation of zoophilic and anthropophilic strains of Trichophyton interdigitale and other species of T. mentagrophytes sensu lato. Br J Dermatol. 2010. 162:282–295.
17. Clinical and Laboratory Standards Institute. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing: approved guideline. CLSI document MM18-A. 2008. Wayne, PA: Clinical and Laboratory Standards Institute.
18. Balajee SA, Gribskov JL, Hanley E, Nickle D, Marr KA. Aspergillus lentulus sp. nov., a new sibling species of A. fumigatus. Eukaryot Cell. 2005. 4:625–632.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download