Abstract
Background
The aims of this study were to compare several DNA extraction methods and 16S rDNA primers and to evaluate the clinical utility of broad-range PCR in continuous ambulatory peritoneal dialysis (CAPD) culture fluids.
Methods
Six type strains were used as model organisms in dilutions from 108 to 100 colony-forming units (CFU)/mL for the evaluation of 5 DNA extraction methods and 5 PCR primer pairs. Broad-range PCR was applied to 100 CAPD culture fluids, and the results were compared with conventional culture results.
Results
There were some differences between the various DNA extraction methods and primer sets with regard to the detection limits. The InstaGene Matrix (Bio-Rad Laboratories, USA) and Exgene Clinic SV kits (GeneAll Biotechnology Co. Ltd, Korea) seem to have higher sensitivities than the others. The results of broad-range PCR were concordant with the results from culture in 97% of all cases (97/100). Two culture-positive cases that were broad-range PCR-negative were identified as Candida albicans, and 1 PCR-positive but culture-negative sample was identified as Bacillus circulans by sequencing. Two samples among 54 broad-range PCR-positive products could not be sequenced.
Although the conventional phenotypic method is still popular to identify common bacteria in clinical microbiology laboratories, it is difficult to use this method when bacteria reveal uncommon phenotypes, grow slowly, or are not included in commercial kit databases [1, 2]. In addition, we cannot detect unculturable or fastidious microorganisms or organisms in patients who have recently received antibiotic therapy, even when bacteria are present in the clinical samples [3, 4].
To overcome these problems, many molecular techniques have been adopted. Broad-range PCR is highly useful for bacterial detection and identification [5, 6]. Broad-range PCR using 16S rDNA is based on the fact that this target gene has highly conserved sequences in most bacterial species [7]. DNA extraction is the first step and is important because the DNA concentrations can differ according to the extraction method used. Thus, the final diagnostic efficiency is influenced by the extraction method. Various methods, including heating and commercial kits, are used in the clinical laboratory, but there are only a few comparison studies of the several nucleic acid extraction kits being used to obtain samples for broad-range PCR [3, 6, 8, 9]. The primer pair used for 16S rDNA is also relevant because the test can reveal different results according to the size and position of the analyzed gene portion [10].
Peritonitis is one of the most common complications of continuous ambulatory peritoneal dialysis (CAPD), and rapid and accurate identification of the causative pathogen is essential for diagnosis and selection of the appropriate therapy [11-14]. However, conventional culture takes at least 2 or 3 days to provide the final identification, and we would be in a difficult situation if there were small numbers of bacteria or fastidious bacteria in the CAPD fluid. It would be valuable to use supplementary molecular methods such as broad-range PCR for detection and identification of pathogens.
The aims of this study were to select the most appropriate DNA extraction method and 16S rDNA primer pair for broad-range PCR by comparing several extraction methods and primer pairs, and to evaluate the clinical utility of broad-range PCR in CAPD culture fluids using the optimal DNA extraction method and primer.
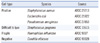
Six type strains were used for the evaluation of DNA extraction methods (Table 1). Among these, Staphylococcus aureus, Escherichia coli, and Pseudomonas aeruginosa were used as positive control strains; Streptococcus pyogenes was used as an example of a strain that is difficult to lyse, and Haemophilus influenzae was used as a representative of fragile bacteria. Candida albicans was used as negative control. All strains were grown on blood and chocolate agar plates for 24 hr and diluted until appropriate concentrations were reached at a McFarland standard of 0.5. Using distilled water, we performed 10-fold serial dilutions from 108 colony-forming units (CFU)/mL to 100 CFU/mL.
All dilutions were centrifuged, and the supernatant was harvested, except when the QIAmp DNA Mini kit (Qiagen GmbH, Hilden, Germany) and the heating method were used, for which we left 200 µL of supernatant on top of the pellets. The commercial DNA extraction kits used in this study were the InstaGene Matrix (Bio-Rad Laboratories, Hercules, CA, USA), Exgene™ Clinic SV kit (GeneAll Biotechnology Co. Ltd., Seoul, Korea), QIAmp DNA Mini kit (Qiagen GmbH), and Easy-DNA™ kit (Invitrogen, Carlsbad, CA, USA). The InstaGene Matrix is composed of 6% InstaGene Matrix and a magnetic stir bar. Boiling in the presence of the matrix resulted in cell lysis. The Exgene™ Clinic SV kit and QIAmp DNA Mini kit utilize the silica-binding technology to purify DNA. The DNA in the lysates binds to the silica membrane, and impurities pass through the membrane into a collection tube. The Easy-DNA kit uses ethanol precipitation purification with chloroform. All procedures were performed according to the manufacturer's instructions. For the heating method, all diluted samples were heated at 100℃ for 10 min and placed on ice for 5 min. After centrifugation, supernatant was used as the source of DNA for PCR analysis. We used the 27F and 515R primer set to compare the DNA extraction methods (Table 2).
We selected 5 primer sets on the basis of previous reports to compare their efficacy for 16S rDNA PCR (Table 2) [15-20]. Each primer set has a different amplification size and position of 16S rDNA. We used InstaGene Matrix to extract DNA for the evaluation of PCR primers on the basis of our study results.
Amplification of bacterial 16S rDNA was carried out with a Veriti™ Thermal Cycler (Applied Biosystems, Singapore, Singapore). The final reaction mixture (25 µL) contained 2 µL of DNA eluate, 10 pM of each primer, 0.85 U of AmpliTaq Gold® LD DNA polymerase (Applied Biosystems, Foster City, CA, USA), 2.0 mL of MgCl2, 0.2 mM dNTP (Applied Biosystems), and 2 µL of 10×PCR buffer. The amplicons were detected by electrophoresis on a 2% agarose gel containing 1 µL of ethidium bromide. All procedures for the comparison of the DNA extraction methods and primer sets were repeated in triplicate.
A total of 100 CAPD culture fluids from blood culture bottles were collected from January to September 2009. We performed all culture procedures according to the 2005 update of the International Society for Peritoneal Dialysis (ISPD) guidelines/recommendations [21]. We employed a BacT/Alert blood culture system (bioMérieux, Durham, NC, USA) with inoculation of the sediment obtained by centrifuging 50 mL of effluent, as recommended by the ISPD guidelines.
Next, 10 µL of CAPD culture fluid was added to 1 mL of autoclaved nanopure water in a 1.5 mL microtube for DNA extraction. We inverted the tube several times and then incubated the sample at room temperature for 30 min. The sample was centrifuged at 13,000 rpm for 2 min, and the supernatant was removed. We used InstaGene Matrix as a DNA extraction method and the primer set of 27F and 515R.
The purified DNA obtained by PCR was sequenced with an ABI Prism 3130xl genetic analyzer (Applied Biosystems) and a BigDye Terminator v 3.1 cycle sequencing kit (Applied Biosystems). The primers used for sequencing were the same as those used for PCR amplification. All sequences were compared with similar sequences of the reference organisms using MicroSeq 500 v 2.0 software.
We also compared the final strain identifications using 16S rDNA sequencing with the culture results based on conventional biochemical tests and the automated Vitek 2 identification system (bioMérieux, St. Louis, MO, USA).
The bacterial detection limits of 5 DNA extraction methods using 5 type strains are shown in Table 3. There were some differences between the DNA extraction methods. By simple heating, all bacterial species could be detected at a concentration of 108 CFU/mL, whereas all commercial kits had lower detection limits. Among the commercial kits, the Exgene™ Clinic SV and InstaGene Matrix had lower detection limits than the QIAamp DNA mini kit and the Easy-DNA™. When using InstaGene Matrix, H. influenzae and P. aeruginosa could be detected at concentrations of 103 CFU/mL and E. coli, S. aureus, and S. pyogenes at 104 CFU/mL. The Easy-DNA™ kit had the highest detection limit. These results suggest that commercial kits are superior to simple heating, but there are some differences among the kits. Three commercial kits showed lower detection limits for H. influenzae than for other bacterial species.
The DNA extraction time was different for each method. Simple heating was the most rapid extraction method, taking about 20 min. For commercial kits, the time needed for DNA extraction was 40 min for InstaGene Matrix and 1 hr for Exgene™ Clinic SV and QIAamp DNA. Using the Easy-DNA™ took the longest time, 2 hr. Therefore, we chose the InstaGene Matrix as the DNA extraction kit for the evaluation of primer pairs and extraction methods for broad-range PCR from CAPD culture fluids.
We compared 5 primer pairs for broad-range PCR using the InstaGene Matrix for DNA extraction. When we used the 27F and 1492R primers, we could detect the full sequence of 16S rDNA at the level of 106 CFU/mL, except in S. pyogenes (107 CFU/mL) (Table 4). The primer pair of 27F and 515R showed the lowest detection limit (103 to 104 CFU/mL) among the 5 pairs. The results with Bak11w and Bak2 were variable in repeated tests. Most primer pairs showed lower detection limits for H. influenzae and there were some differences in the detection limit with regard to the bacterial species.
On the basis of our study results, we selected InstaGene Matrix and the 27F/515R primer pair as DNA extraction method and primer pair, respectively, for broad-range PCR. Among 100 CAPD culture fluids, 55 specimens were positive in culture. Among the 55 culture-positive specimens, 53 also were positive by PCR analysis, and 1 specimen showed a positive result by broad-range PCR, although the culture was negative (Table 5). Two specimens that were culture-positive but PCR-negative were finally identified as containing C. albicans. One specimen that was PCR-positive but culture-negative was identified as containing Bacillus circulans by 16S rDNA sequencing. Two strains could not be appropriately sequenced, although these were positive by broad-range PCR.
On the basis of the final identifications, coagulase-negative staphylococci (CoNS) (N=19) were the most common pathogen, followed by E. coli (N=8), Bacillus species (N=8), Streptococcus species (N=6), S. aureus (N=4), Micrococcus species (N=2), and others (N=5).
Thirty-three strains were identified to the species level by 16S rDNA sequencing when we used the CLSI MM-18A guideline (Table 6). The 16S rDNA sequencing using broad-range PCR could identify 2 Micrococcus, 6 Bacillus, and 1 Gornodia to the species level, although these could not be identified by conventional biochemical methods with commercial identification kits.
Eleven strains were identified only to the genus level by the CLSI MM-18A guideline. Two strains showed lower than 99% similarity with type strains, and 9 of the 11 strains were not identified to the species level because the difference between the first and second matched sequences was less than 0.8% (0.0% to 0.41%) (Table 7). The 8 E. coli strains could not be differentiated from Shigella using 16S rDNA sequencing, even though they could be easily distinguished by conventional methods.
The use of molecular methods is increasing in clinical laboratories to overcome the limitations of conventional culture methods. Of these methods, broad-range PCR and sequencing are attractive because by using them, most bacteria can be detected, regardless of their species and specific culture conditions [5, 6]. We could detect and identify bacterial pathogens directly from CAPD culture fluids by application of broad-range PCR using 16S rDNA.
Extraction of DNA from clinical specimens is an important step in obtaining a reliable result. Some bacteria, such as gram-positive organisms and mycobacteria, are difficult to lyse because of the strength of their cell walls, emphasizing the importance of DNA extraction [3]. The optimal extraction method for broad-range PCR should concentrate DNA from target bacteria and remove inhibitory factors commonly present in specimens and culture fluids. A simple handling process is also highly desirable [3]. The phenol-chloroform method was commonly used in the past but is not as popular today because phenol has some undesirable features, including corrosiveness and toxicity [22]. The use of commercial DNA extraction kits is rapidly increasing in clinical laboratories because they are convenient, easy to use, and offer valid test results.
We compared 5 DNA extraction methods, including 4 commercial kits, to obtain material for broad-range PCR. All 4 commercial kits were superior to simple heating. There were some differences in the detection limits of the various methods. In particular, the InstaGene Matrix and Exgene™ Clinic SV kits showed lower detection limits than the QIAmp DNA mini kit and Easy-DNA™ kit. The detection limits of the InstaGene Matrix ranged from 103 to 104 CFU/mL, but those of the Easy-DNA™ kit were 107 CFU/mL for all 5 bacterial species. Differences among DNA extraction methods have been documented in previous reports [3, 9]. In the study by Rantakokko-Jalava et al. [3], the range of the detection limits of 5 commercial nucleic acid extraction kits was 103 to >105 CFU/mL. Their results were also different according to the kits used. The detection limits of the Masterpure DNA purification kit were 103 to 104 CFU/mL, whereas those of the High Pure PCR template preparation kit were 104 to >105 CFU/mL. Although the kits were different, the results of our study were similar to those of an earlier study. Zucol et al. [9] compared 3 DNA extraction protocols for broad-range real-time PCR assays, and the range of the detection limits was 1 to >106 CFU/mL. They used 3 modified protocols with the QIAmp DNA blood mini kit and Wizard SV genomic DNA purification system. These methods also showed differences in detection limits according to the DNA extraction protocol used. These differences could affect test sensitivity and specificity; thus, it is imperative to select an appropriate DNA extraction method to obtain a reliable result. We also analyzed the processing time and costs for each method. Simple heating was the most rapid extraction method, taking about 20 min, whereas the processing time of the 4 commercial kits ranged from 40 min to 2 hr. Among these kits, the InstaGene Matrix kit took the least time. The costs of commercial kits varied. The QIAmp DNA mini kit and Easy-DNA™ kit were more expensive than the Exgene™ Clinic SV kit and InstaGene Matrix kit. On the basis of our results, we used InstaGen Matrix for the evaluation of primers and broad-range PCR for CAPD culture fluids.
The primers for broad-range PCR using 16S rDNA can also affect the amplification results. Several primer pairs have been used in previous studies [15-20, 23]; thus, the investigators used different positions and sizes of 16S rDNA. We confirmed some differences in the detection limits according to the primer sets. The 27F/515R set showed the lowest detection limit (range 103 to 104 CFU/mL). In the study by Zucol et al. who used the same DNA extraction protocols, the detection limits of the Bak11W/Bak2 primer set were between 1 and 103 CFU/mL; however, those of the 16SFa/16SFb/16SR primer set were between 102 and 106 CFU/mL [9]. Therefore, the influence of the primer set was also an important factor for the determination of detection limits. We repeated broad-range PCR in triplicate to calculate the reproducibility for each primer set, and most primer sets showed the same results each time, although the results of the Bak11w/Bak2 primer set varied. Finally, we selected the 27F/515R primer pair and InstaGene Matrix for the detection and identification of bacteria in CAPD culture fluids. We used broad-range PCR to detect various bacterial species, not a specific species. However, the detection limits were different according to the species in our study and that of Zucol et al. [9], which affects the sensitivity of broad-range PCR.
We compared broad-range PCR followed by sequencing to the culture results. We could detect 53 of 55 culture-positive CAPD culture fluids. The remaining 2 samples, showing culture-positive, PCR-negative results, yielded C. albicans. This result is appropriate because the broad-range PCR for 16S rDNA can detect only bacteria; therefore, this test should be used as a supplement to conventional culture methods. One sample showing a distinctive result (culture-negative but PCR-positive) was identified as containing a B. circulans, because sequencing revealed 100% similarity to a reference strain. Although we could not determine where this discrepancy originated from, it could be due to the absence of growth on subculture on media, lower numbers of bacteria, and remnants of bacterial DNA in sterilized culture bottles. In total, 33 and 44 strains were correctly identified to the species and genus level, respectively, using broad-range PCR and sequencing. The 16s rDNA sequencing using broad-range PCR was valuable in identifying gram-positive bacilli. In general, it is difficult to identify these bacteria using conventional biochemical methods with commercial kits because there are limited databases and kits in the commercial identification systems. We also identified 2 micrococci to the species level, whereas the biochemical methods did not reach this level. We can make good use of broad-range PCR directly on CAPD culture fluids for accurate identification of bacteria. However, 16S rDNA sequencing could not differentiate Escherichia coli and Shigella. In addition, Enterococcus faecium/Enterococcus hirae, Staphylococcus capitis/Staphylococcus caprae, and Streptococcus salivarius/Streptococcus vestibularis were not differentiated by 16S rDNA sequencing. The reason for this is that these strains are genetically closely related. We could not obtain sequencing results in 2 specimens, although the broad-range PCR performance was good. These specimens showed multiple peaks and very short sequences; thus, we assumed presence of 2 or more bacteria and contamination of the clinical specimens by environmental organisms. The limitation of this technique in species identification needs to be considered and, consequently, this method should be used as a supplement to culture methods.
There are a few limitations of this study. We used the 0.5 McFarland standard with a photometric device for inoculum preparation. We considered the samples to contain 1×108 CFU/mL, but this is only an approximation. Therefore, it is possible that we miscalculated the detection limits of each DNA extraction method and each primer set. In addition, we used only 1 primer set, 27F and 515R, to evaluate 5 DNA extraction methods. In a previous report [9], it was suggested that combination of the DNA extraction protocol and primer pair determines analytical sensitivity. This suggests that the results might have been different if we had used all 5 primer sets to evaluate each extraction method. The presence of PCR inhibitors may affect the sensitivity of various extraction methods, but we did not evaluate their effects in this study. We should note that reaction inhibitors could influence most PCR assays, especially when clinical specimens are used. An additional limitation of this study is that no statistical analysis was performed for comparison of detection limits of the extraction methods and primer sets; therefore, the results are limited in their suitability for objective interpretation. Consequently, it is necessary to evaluate their performance before use in each clinical laboratory to optimize the efficiency, considering the nucleic acid extraction method, primer set, PCR inhibitors, and other factors.
In conclusion, we demonstrated differences in the analytical sensitivity of various DNA extraction methods and broad-range PCR primers. We showed that broad-range PCR could be used to detect bacterial pathogens directly in CAPD culture fluid as a supplement to culture methods for the diagnosis of peritonitis caused by CAPD.
Figures and Tables
References
1. Petti CA, Polage CR, Schreckenberger P. The role of 16S rRNA gene sequencing in identification of microorganisms misidentified by conventional methods. J Clin Microbiol. 2005. 43:6123–6125.

2. Shin JH, Kim HR, Lee JN. Clinical significance and species identification of rapidly growing mycobacteria isolated from routine blood cultures. Korean J Lab Med. 2005. 25:162–167.
3. Rantakokko-Jalava K, Jalava J. Optimal DNA isolation method for detection of bacteria in clinical specimens by broad-range PCR. J Clin Microbiol. 2002. 40:4211–4217.

4. Yoo TH, Chang KH, Ryu DR, Kim JS, Choi HY, Park HC, et al. Usefulness of 23S rRNA amplification by PCR in the detection of bacteria in CAPD peritonitis. Am J Nephrol. 2006. 26:115–120.

5. Sontakke S, Cadenas MB, Maggi RG, Diniz PP, Breitschwerdt EB. Use of broad range16S rDNA PCR in clinical microbiology. J Microbiol Methods. 2009. 76:217–225.

6. Fenollar F, Lévy PY, Raoult D. Usefulness of broad-range PCR for the diagnosis of osteoarticular infections. Curr Opin Rheumatol. 2008. 20:463–470.

7. Drancourt M, Bollet C, Carlioz A, Martelin R, Gayral JP, Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000. 38:3623–3630.

8. Fuursted K, Arpi M, Lindblad BE, Pedersen LN. Broad-range PCR as a supplement to culture for detection of bacterial pathogens in patients with a clinically diagnosed spinal infection. Scand J Infect Dis. 2008. 40:772–777.

9. Zucol F, Ammann RA, Berger C, Aebi C, Altwegg M, Niggli FK, et al. Real-time quantitative broad-range PCR assay for detection of the 16S rRNA gene followed by sequencing for species identification. J Clin Microbiol. 2006. 44:2750–2759.

10. Maiwald M. David H, editor. Broad-range PCR for detection and identification of bacteria. Molecular Microbiology: Diagnsotic principles and practice. 2004. Washington D.C.: ASM Press;379–390.
11. Piraino B. Peritonitis as a complication of peritoneal dialysis. J Am Soc Nephrol. 1998. 9:1956–1964.

12. Kim DK, Yoo TH, Ryu DR, Xu ZG, Kim HJ, Choi KH, et al. Changes in causative organisms and their antimicrobial susceptibilities in CAPD peritonitis: a single center's experience over one decade. Perit Dial Int. 2004. 24:424–432.

13. Zelenitsky S, Barns L, Findlay I, Alfa M, Ariano R, Fine A, et al. Analysis of microbiological trends in peritoneal dialysis-related peritonitis from 1991 to 1998. Am J Kidney Dis. 2000. 36:1009–1013.

14. Lee JY, Kim SH, Jeong HS, Oh SH, Kim HR, Kim YH, et al. Two cases of peritonitis caused by Kocuria marina in patients undergoing continuous ambulatory peritoneal dialysis. J Clin Microbiol. 2009. 47:3376–3378.
15. Goldenberger D, Künzli A, Vogt P, Zbinden R, Altwegg M. Molecular diagnosis of bacterial endocarditis by broad-range PCR amplification and direct sequencing. J Clin Microbiol. 1997. 35:2733–2739.

16. Mignard S, Flandrois JP. 16S rRNA sequencing in routine bacterial identification: a 30-month experiment. J Microbiol Methods. 2006. 67:574–581.

17. Edwards U, Rogall T, Blöcker H, Emde M, Böttger EC. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989. 17:7843–7853.

18. Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985. 82:6955–6959.

19. Rovery C, Greub G, Lepidi H, Casalta JP, Habib G, Collart F, et al. PCR detection of bacteria on cardiac valves of patients with treated bacterial endocarditis. J Clin Microbiol. 2005. 43:163–167.

20. Eden PA, Schmidt TM, Blakemore RP, Pace NR. Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction-amplified 16S rRNA-specific DNA. Int J Syst Bacteriol. 1991. 41:324–325.
21. Piraino B, Bailie GR, Bernardini J, Boeschoten E, Gupta A, Holmes C, et al. Peritoneal dialysis-related infections recommendations: 2005 update. Perit Dial Int. 2005. 25:107–131.

22. Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987. 162:156–159.

23. Kemp M, Holtz K, Andresen K, Christensen JJ. Demonstration by PCR and DNA sequencing of Corynebacterium pseudodiphtheriticum as a cause of joint infection and isolation of the same organism from a surface swab specimen from the patient. J Med Microbiol. 2005. 54:689–691.




 PDF
PDF ePub
ePub Citation
Citation Print
Print







 XML Download
XML Download