Abstract
Objective
To evaluate the performance of Momguard, non-invasive prenatal test (NIPT) for detecting trisomy (T) 21, T18, T13, and sex-chromosome abnormalities recently developed in Korea.
Methods
This preliminary study formed part of a large prospective cohort study conducted at Asan Medical Center, Seoul, Korea. Only pregnant women who underwent both NIPT and confirmatory karyotyping were included in this study. NIPT results were compared with those of karyotype analyses.
Results
Among 93 eligible cases, NIPT results could not be obtained in one case due to a low fetal cell-free DNA fraction. Based on NIPT, eight cases of fetal aneuploidies, including T21 (n=5), T18 (n=2), and T13 (n=1), were identified. For T21 and T18, the sensitivity and specificity of NIPT were both 100%, with a false-positive and false-negative rate of 0% and a positive-predictive value of 100%. One patient classified as having intermediate risk for T13 by NIPT was confirmed to have T13 by karyotyping, and there were no false-negative cases. No cases of sex-chromosome anomalies were detected by NIPT or karyotyping during the study period.
Conventional prenatal testing for the diagnosis of fetal chromosomal aneuploidies includes non-invasive screening using maternal serum and ultrasound scanning, or invasive diagnostic procedures, such as chorionic villus sampling (CVS) or amniocentesis. Although invasive tests are highly accurate, they carry risks of procedure-related miscarriage [12]. To develop a prenatal diagnostic procedure that carries no risk of abortion, various non-invasive screening methods have been introduced. Methods based on maternal serum biochemical markers and ultrasound scanning have improved markedly in recent years. Although fully integrated screening tests are safe, they have a 2 to 3% false-positive rate (FPR) and a ≥5% false-negative rate (FNR) for detecting trisomy (T) 21 and T18 [345], and are less effective for detecting aneuploidies other than T21 or T18. Therefore, they induce significant maternal anxiety and unnecessary fetal loss due to subsequent invasive diagnostic procedures.
In 1997, Lo et al [6] first reported the presence of fetal cell-free DNA (cfDNA) in maternal plasma during pregnancy. Since then, there have been many attempts to develop non-invasive prenatal testing (NIPT) methods for detecting fetal gender, fetal RhD genotype, paternally inherited or de novo mutations, and monogenetic disorders [7]. Early assays were based on sensitive polymerase chain reaction (PCR)-based amplification of unique sequences from maternal plasma. However, most PCR-based assays are limited by PCR primer specificity and assay sensitivity. Moreover, 99% homology between maternal and cfDNA complicates detection of fetal aneuploidy using traditional PCR technology [78].
The recent rapid development of next generation sequencing (NGS) and powerful bioinformatic technologies have initiated a new era of prenatal testing for the evaluation of fetal aneuploidies, especially T21, T18, and T13. Various NGS-based NIPT methods have been developed with detection sensitivity exceeding 99% for T21 and T18, with a FPR of less than 1% [9101112]. A similar accuracy has been achieved for T13 detection, with sensitivity ranging from 78.7% to 100% and a FPR of <1% [71314].
Since its initial development in Hong Kong in 2011, commercial NIPT has spread rapidly worldwide. Although clinicians in Korea have adopted NIPT as avidly as clinicians in other countries, the tests are not yet performed locally: samples collected in Korea are generally shipped abroad for processing in overseas laboratories. Moreover, NIPT has not been clinically evaluated using a Korean cohort.
Momguard is an independent NIPT protocol based on random massively parallel shotgun sequencing and was recently developed in Korea. A prospective, large-scale blinded clinical study for evaluating its utility is currently underway. In this pilot study, we evaluated the performance of Momguard in a preliminary cohort.
The clinical evaluation of Momguard was conducted as a blinded prospective cohort study at Asan Medical Center from August 2014, with the goal of reaching 1,000 participants. Approval was obtained from our institutional review board and written informed consent was obtained from all participants. The study included pregnant women who were >18 years old, gestational age >8 weeks, and who met at least one of the following additional criteria: advanced maternal age (≥35 years), a positive serum biochemical screening test, the presence of fetal anomalies detected by ultrasound, or a personal/family history of fetal aneuploidy. To evaluate the performance of Momguard in twin pregnancies, multiple gestations were also included.
For this preliminary study, part of the clinical data collected between August 2014 and February 2015 was analyzed. This analysis included all pregnant women who underwent either CVS, amniocentesis, or cordocentesis for confirming fetal karyotype. Fetuses with karyotypes that were confirmed using peripheral blood after birth or conceptual tissues in cases of missed abortion were also included. In cases of cardiac malformation or a suspected genetic syndrome detected by ultrasonography, fluorescence in situ hybridization was also performed to detect microdeletions.
Maternal whole blood samples (10 mL) were collected in cell-free DNA BCT tubes (Streck, Omaha, NE, USA). Most samples arrived at the laboratory without processing within 24 hours of collection. Plasma was isolated from the maternal blood by two-step centrifugation at 1,600 ×g for 10 minutes and then at 16,000 ×g for 10 minutes at 4℃. The isolated plasma was distributed into 1 mL aliquots in separate 1.5 mL tubes that were labeled with a distinctive sample code and frozen at -80℃ until required for analysis. All clinical data, including invasive prenatal test results, were blinded to the laboratory investigators.
cfDNA was isolated from 2 mL of maternal plasma using a QIAmp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions. The extracted cfDNA was eluted in 40 µL of AVE buffer (Qiagen). A cfDNA library was generated following the Momguard library preparation protocol and was evaluated using a PicoGreen assay and the 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA).
Equal volumes of the resulting libraries were pooled, in a flowcell, for up to 12 samples (MiSeq, Illumina, San Diego, CA, USA) or 96 samples (NextSeq, Illumina), for cluster generation. The pooled libraries were sequenced on MiSeq and NextSeq instruments with MiSeq v3 and NextSeq v2 sequencing chemistry, respectively.
NGS data analyses were performed using our own bioinformatics pipeline. Briefly, the paired-end sequencing reads of 75 bases in length were binned for each sample, according to the index sequence, and mapped to the human genome assembly (hg19). BWA was used to align the sequences to the human genome, and the number of reads that were uniquely aligned to each chromosome was counted. The relative ratio of chromosomes 21, 18 and 13 to autosomes was calculated for each sample. The proportions of reads from sex chromosomes were calculated and used to estimate the fetal DNA fractions in maternal plasma for pregnancies involving male fetuses. Briefly, a Z-score was derived for a test sample by subtracting the residuals of the ratio of the relevant chromosome to the autosomes for a set of euploid pregnancies by those for the standard set, and dividing this by the standard deviation of the residuals for the standard set. Generally, a high Z-score (i.e., >4) for a chromosome indicates that the risk of aneuploidy is higher than that for the standard set. Patients with Z-scores between 2.5 and 4 were considered to exhibit an intermediate risk for T21 and T18. Different criteria were applied for T13; a Z-score of 2.8 was the threshold for high risk, and 1.9 was the cutoff for intermediate risk.
Maternal and gestational ages were expressed as medians and ranges. The distribution of gestational ages and indicated pregnancies were presented as percentages. The coefficients and P-value for the relationship between the cfDNA fraction and gestational age were determined by linear regression analysis. Statistical significance was defined as P<0.05.
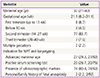
Among 93 pregnant women who underwent both NIPT and karyotyping during the study period, NIPT sequencing was impossible in one case (sampling performed at 8.6 weeks of gestation) owing to an insufficient fraction of cfDNA (<0.5%). Therefore, 92 pregnant women were eligible for this preliminary study; demographic characteristics of the subjects are shown in Table 1. Four pregnant women were karyotyped before 10 weeks of gestation due to recurrent abortion. The cfDNA fraction was sufficient for sequence analysis in these four cases and there was a trend toward a higher cfDNA fraction with advanced gestational age (Fig. 1).
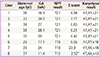
Based on cfDNA analysis, eight cases of high or intermediate risk for fetal aneuploidies, including T21 (n=5), T18 (n=2), and T13 (n=1), were identified (Fig. 2) and these results based on sequencing data were compared with karyotype results (Table 2). The five positive NIPT results for T21 were all confirmed by karyotyping; therefore, the sensitivity was 100% (95% confidence interval [CI], 47.95% to 100%), specificity was 100% (95% CI, 95.8% to 100%), FPR was 0%, FNR was 0%, and the positive-predictive value (PPV) was 100%. For T18, the sensitivity and specificity were also 100% (95% CI, 19.29% to 100% and 95.94% to 100%, respectively), the FPR and FNR were 0%, and PPV was 100%. For T13, only one case (case 8) exhibited intermediate risk based on NIPT, and this was confirmed to have T13 by karyotyping. No false-negative cases were observed. During the study period, no sex-chromosome anomalies were detected by NIPT or karyotyping in this cohort.
Apart from T21, T18, and T13, an additional 10 chromosome abnormalities were detected by cytogenetic analysis with fluorescence in situ hybridization, including one case with triploidy (69, XXX), one with T16, two with derivative chromosomes, five with a 22q11.2 microdeletion, and one with a 17p13.3 microdeletion, none of which were detectable using Momguard.
This pilot study demonstrated that Momguard is a highly accurate prenatal testing tool for the detection of T21 and T18 and did not yield false positives or false negatives. All cfDNA analyses were performed in the LabGenomics Clinical Laboratory (Seongnam, Korea), where a multi-platform NGS-based noninvasive test was implemented for fetal aneuploidy screening. T21 was detected using the Illumina MiSeq and NextSeq 500 NGS platforms, and the results obtained using the two platforms correlated well (data not shown). These results indicated that both sequencing platforms can be used to detect trisomies other than T21 using the standard rules for determining aneuploidy, although the platforms differed in throughput. Only one case failed owing to an insufficient cfDNA fraction. To our knowledge, this is the first clinical study evaluating an independent NIPT protocol using a Korean cohort and confirmed the effectiveness of using Momguard to detect T21 and T18.
It was difficult to assess the clinical usefulness of Momguard for detecting T13 owing to its rarity. One sample classified as intermediate risk was identified in the cfDNA analysis, and T13 was then verified by CVS. To detect T13, we applied a different Z-score threshold, which was empirically derived. The detection rate for T13 is known to be lower than that of T21 (91.6%) and it has a higher FPR (0.097%) [14]. Less accurate NIPT results for T13 have been attributed to differences in the guanine and cytosine content, which is lower, on average, for T13 than T21 [16]. Therefore, although NIPT may be considered as an alternative to invasive testing, its application in the diagnosis of T13 is still limited. Most fetuses with T13 have multiple anomalies, and therefore, detailed ultrasonographic evaluation in addition to NIPT would increase the diagnostic accuracy. Patient enrollment is currently in progress for a large prospective study for evaluating the use of Momguard, and its accuracy for T13 detection will require further evaluation.
The fetal cfDNA fraction varies among pregnant individuals and gestational ages, but its median is 10.2% between 10 and 21 weeks of gestation and it increases by >1% every week [17]. Our preliminary results also demonstrated an increase in the cfDNA fraction with advanced gestational age (Fig. 1). Recently, Song et al reported that NIPT can be used between 8 and 12 weeks, with the median cfDNA fraction of 8.54% [18]. We successfully analyzed the cfDNA of four samples obtained before 10 weeks, and the cfDNA fractions were 10.8% at 8.2 weeks, 4.03% at 8.4 weeks, 4.31% at 9 weeks, and 6.6% at 9.4 weeks, respectively. We also successfully evaluated one case wherein a relatively low fetal fraction (3.85%) was observed; sampling had been performed at 11.4 weeks (shown in Fig. 1).
Currently, NIPT is available for T21, T18, T13, and sex-chromosome abnormalities. Although NIPT is now accepted as a highly reliable and accurate method for detecting T21 and T18 (with both sensitivity and specificity >90%), some false-positive cases are consistently reported. Therefore, it cannot yet be considered as a diagnostic test, and confirmative karyotyping should be offered to women with positive NIPT results. In this preliminary study, the clinical usefulness of Momguard was only assessed with respect to T21, T18, and T13, and its applications to other cytogenetic anomalies are currently limited. However, rapid improvements in sequencing technology may enable the detection of several additional cytogenetic abnormalities. Development of the next version of Momguard, which will make screening for several microdeletion syndromes possible, is in progress.
Based on this preliminary study, we confirmed the utility of Momguard, and demonstrated its excellent reliability in the detection of T21 and T18. Further prospective studies should evaluate the performance of Momguard for detecting T13 and sex-chromosome anomalies.
Figures and Tables
Fig. 1
Correlation between cell-free DNA (cfDNA) fraction and gestational age in 46 pregnancies with male fetuses (R2=0.1636, P<0.01). Unlike for male fetuses, for which Y chromosomes are effective markers, it is particularly difficult to quantitate the fetal fraction in female fetuses; there are no universal and reliable fetal markers available to estimate the fetal fraction in maternal plasma [15]. Therefore, cfDNA level was analyzed using only male fetuses.
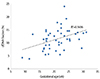
Fig. 2
Interactive dot diagram for fetal aneuploidy. The Z-scores of chromosomes 21, 18 and 13 are represented on the Y-axis. The solid line represents the high risk cutoff value, and the dotted line represents the intermediate risk cutoff value for each aneuploidy test. T, trisomy.

Acknowledgments
This study was supported by a grant from the LabGenomics Clinical Research Institute.
References
1. American College of Obstetricians and Gynecologists. ACOG Practice Bulletin No. 88, December 2007. Invasive prenatal testing for aneuploidy. Obstet Gynecol. 2007; 110:1459–1467.
2. Tabor A, Alfirevic Z. Update on procedure-related risks for prenatal diagnosis techniques. Fetal Diagn Ther. 2010; 27:1–7.
3. Malone FD, Canick JA, Ball RH, Nyberg DA, Comstock CH, Bukowski R, et al. First-trimester or second-trimester screening, or both, for Down's syndrome. N Engl J Med. 2005; 353:2001–2011.
4. Nicolaides KH. Nuchal translucency and other first-trimester sonographic markers of chromosomal abnormalities. Am J Obstet Gynecol. 2004; 191:45–67.
5. Rozenberg P, Bussieres L, Chevret S, Bernard JP, Malagrida L, Cuckle H, et al. Screening for Down syndrome using first-trimester combined screening followed by second-trimester ultrasound examination in an unselected population. Am J Obstet Gynecol. 2006; 195:1379–1387.
6. Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997; 350:485–487.
7. Swanson A, Sehnert AJ, Bhatt S. Non-invasive prenatal testing: technologies, clinical assays and implementation strategies for women's healthcare practitioners. Curr Genet Med Rep. 2013; 1:113–121.
8. Lo YM, Chan KC, Sun H, Chen EZ, Jiang P, Lun FM, et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med. 2010; 2:61ra91.
9. Benn P, Cuckle H, Pergament E. Non-invasive prenatal testing for aneuploidy: current status and future prospects. Ultrasound Obstet Gynecol. 2013; 42:15–33.
10. Gregg AR, Van den Veyver IB, Gross SJ, Madankumar R, Rink BD, Norton ME. Noninvasive prenatal screening by next-generation sequencing. Annu Rev Genomics Hum Genet. 2014; 15:327–347.
11. Ehrich M, Deciu C, Zwiefelhofer T, Tynan JA, Cagasan L, Tim R, et al. Noninvasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: a study in a clinical setting. Am J Obstet Gynecol. 2011; 204:205.e1–205.e11.
12. Norton ME, Brar H, Weiss J, Karimi A, Laurent LC, Caughey AB, et al. Non-Invasive Chromosomal Evaluation (NICE) Study: results of a multicenter prospective cohort study for detection of fetal trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012; 207:137.e1–137.e8.
13. Agarwal A, Sayres LC, Cho MK, Cook-Deegan R, Chandrasekharan S. Commercial landscape of noninvasive prenatal testing in the United States. Prenat Diagn. 2013; 33:521–531.
14. Verweij EJ, de Boer MA, Oepkes D. Non-invasive prenatal testing for trisomy 13: more harm than good? Ultrasound Obstet Gynecol. 2014; 44:112–114.
15. Daley R, Hill M, Chitty LS. Non-invasive prenatal diagnosis: progress and potential. Arch Dis Child Fetal Neonatal Ed. 2014; 99:F426–F430.
16. Chiu RW, Chan KC, Gao Y, Lau VY, Zheng W, Leung TY, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci U S A. 2008; 105:20458–20463.
17. Wang E, Batey A, Struble C, Musci T, Song K, Oliphant A. Gestational age and maternal weight effects on fetal cell-free DNA in maternal plasma. Prenat Diagn. 2013; 33:662–666.
18. Song Y, Huang S, Zhou X, Jiang Y, Qi Q, Bian X, et al. Non-invasive prenatal testing for fetal aneuploidies in the first trimester of pregnancy. Ultrasound Obstet Gynecol. 2015; 45:55–60.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download