Abstract
Objective
This study was to identify small inhibitory RNAs (siRNAs) that are effective in inhibiting growth of cervical cancer cell lines harboring human papilloma virus (HPV) and to examine how siRNAs interact with interferon beta (IFN-β) and thimerosal.
Methods
The HPV18-positive HeLa and C-4I cell lines were used. Four types of siRNAs were designed according to their target (both E6 and E7 vs. E6 only) and sizes (21- vs. 27-nucleotides); Ex-18E6/21, Ex-18E6/27, Sp-18E6/21, and Sp-18E6/27. Each siRNA-transfected cells were cultured with or without IFN-b and thimerosal and their viability was measured.
Results
The viabilities of HPV18-positive tumor cells were reduced by 21- and 27-nucleotide siRNAs in proportion to the siRNA concentrations. Of the two types of siRNAs, the 27-nucleotide siRNA constructs showed greater inhibitory efficacy. Sp-18E6 siRNAs, which selectively downregulates E6 protein only, were more effective than the E6- and E7-targeting Ex-18E6 siRNAs. siRNAs and IFN-β showed the synergistic effect to inhibit HeLa cell survival and the effect was proportional to both siRNA and IFN-β concentrations. Thimerosal in the presence of siRNA exerted a dose-dependent inhibition of C-4I cell survival. Finally, co-treatment with siRNA, IFN-β, and thimerosal induced the most profound decrease in the viability of both cell lines.
Conclusion
Long (27-nucleotides) siRNAs targeting E6-E7 mRNAs effectively reduce the viability of HPV18-positive cervical cancer cells and show the synergistic effect in combination with IFN-b and thimerosal. It is necessary to find the rational design of siRNAs and effective co-factors to eradicate particular cervical cancer.
Human papilloma viruses (HPVs) are the etiological agents of cervical and other anogenital malignancies. Over 100 different types of HPVs have been identified to date, and all target epithelial tissues for infection. High-risk group HPVs is associated with the vast majority (99.7%) of patients with uterine cervical cancer, whereas low-risk group HPVs is a cause of genital warts as well as abnormal vaginal smear results [1]. HPVs infect metaplastic cells between the basal cells and the squamocolumnar cells in the cervix and can also infect glandular epithelial cells in the endocervix. Although HPVs exist in episomal forms, they become integrated into host cell DNA, thus increasing the opportunities for cell proliferation and deterioration in high-grade lesions [2]. If this condition persists for several years, transformation into tumor cells occurs. Khan et al. [3] reported that cervical intraepithelial neoplasia grade 3 or cervical cancer occurred in 20% and 15% of HPV16- and HPV18-positive females, respectively, within 120 months of the initial infection.
The HPV genome contains genetic information for at least six early proteins (E1, E2, E4-E7) necessary to reproduce their own DNAs in infected cells. E6 and E7 proteins are very important in the viral reproduction and interaction with host proteins. E7 proteins promote cell-cycle progression from G1 to S and this abnormality in cell differentiation is activated by E6 protein, as a result, cooperative actions of E6 and E7 proteins induce the immortality of host cells [4]. For this reason, these proteins have become primary targets of genetic treatments of HPV-positive cervical cancers. A lot of studies have been conducted to treat HPV infection; however, we have not still solved this problem effectively.
RNA interference (RNAi) is a method for inhibiting gene expression that relies on sequence homology between small double-stranded RNA (dsRNA) molecules. Small interfering RNAs (siRNAs), consisting of 21-25 nucleotides that inhibit posttranscriptional gene expression [5], were discovered in 2001 and have since been widely used as a means for downregulating specific targets. This can be applied to the development of drugs that target genes responsible for human diseases. However, diverse siRNAs exist for the same gene, and individual siRNAs exhibit wide variations in their ability to inhibit gene expression [6].
In this study, we applied RNAi technology to eradicate HPV infection in HPV18-positive cervical cancer cell lines. We also analyzed the interactions between siRNAs and the known cancer-inhibiting agents, interferon beta (IFN-β) and thimerosal (ethylmercurithiosalicylate), in relation to the viability of cancer cells.
HPV18 genome-positive HeLa and C-4I cells, which are cell lines derived from human cervical cancers, were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) and sub-cultured. The culture medium and cell culture reagents used were from GIBCO-BRL Life Technologies (Gaithersburg, MD, USA). Both cell lines were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, antibiotics (100 U/mL penicillin-G and 100 U/mL streptomycin), and GlutaMAX (100 U/mL) at 37℃ in a humidified 5% CO2 incubator.
Human IFN-β cDNAs were ligated into the corresponding sites of the pLEGFP-C1 vector (Clontech, Mountain View, CA, USA). After plating transformed bacteria, plasmid DNA from several randomly selected colonies was isolated. The resulting pLEGFPC1/hIFN-β expression plasmid was transfected into HEK293 (human embryonic kidney cell line) using lipofectamine (Invitrogen, Carlsbad, CA, USA) and stably transfected clones were selected. The supernatant of clones was collected, and IFN-β was obtained using anti-proliferation assays with the human breast cancer cell line, MDA-MB-468.
To inhibit the expression of both E6 and E7 proteins, we transfected 21-nucleotide (Ex-18E6/21) or 27-nucleotide (Ex-18E6/27) versions of E6-E7-specific siRNAs to HeLa and C-4I cells. The target areas of Ex-18E6 siRNA are common to both E6 and E7 mRNAs and E6*I-E7 mRNA splice variants. Effective inhibition of E6 only was achieved using 21-nucleotide (Sp-18E6/21) and 27-nucleotide (Sp-18E6/27) versions of Sp-18E6 siRNA, which targets a boundary area between the E6* splicing donor site and the intron. The following HPV E6 siRNAs, synthesized by Samchully Pharmaceutical Co. (Gwacheon, Korea), were used in this study: (1) Ex-18E6/21 siRNAs, (sense) 5'-CCU GUG UAU AUU GCA AGAC dTdT-3' and (antisense) 5'-GUC UUG CAA UAU ACA CAG GdTdT-3'; (2) Sp-18E6/21 siRNAs, (sense) 5'-GAG GUA UUU GAA UUU GCA UdTdT-3 and (antisense) 5'-AUG CAA AUU CAA AUA CCU CdTdT-3'; (3) Ex-18E6/27 siRNAs, (sense) 5'-CCU GUG UAU AUU GCA AGA CUU AAA GdTdT-3' and (antisense) 5'-CUU UAA GUC UUG CAA UAU ACA CAG GdTdT-3'; (4) Sp-18E6/27 siRNAs, (sense) 5'-GAG GUA UUU GAA UUU GCA UUU AAA GdTdT-3 and (antisense) 5'-CUU UAA AUG CAA AUU CAA AUA CCU CdTdT-3'. The siRNAs targeting GFP (green fluorescent protein) from IDT Co. (Coralville, IA, USA) were used as control. Sense and antisense RNA oligonucleotides were dissolved in 400 mL of RNase-free water, and annealed by incubating at 30℃ for 1 hour. Suspensions of HeLa and C-4I cells, prepared using 1X trypsin-EDTA, were plated in 24-well plates and transfected with mixtures of E6 siRNA (0, 1, 5, 10, 25, 50 nM) and lipofectamine. Human IFN-β (0, 1, 2, 5, 20 mM) and/or thimerosal (0, 0.2, 0.5 mM) (Sigma Chemicals, Perth, WA, USA) were added to siRNA-transfected or -untransfected cells.
After culturing siRNA-transfected (or untransfected) HeLa and C-4I cells with or without thimerosal and/or human IFN-β for 3 days, supernatants were removed and the wells were washed. For crystal violet assays, 100 mL of 0.5% crystal violet (Sigma Chemicals) solution was added to each well, and then they were incubated for 20 minutes at room temperature. After lysing cells with 100% methanol, optical density was measured at a wavelength of 570 nm using a microplate reader (Molecular Devices, Sunnyvale, CA, USA).
Each siRNA (0, 1, 5, 10, 25, 50 nM) induced a concentration-dependent decrease in the viability of HeLa cells and C-4I cells. The differences in viabilities were appearing from the siRNA concentration of 5 nM and were clearly observed at 50 nM. In HeLa cells, the cell viabilities at siRNA concentration of 50 nM were 44.5% for Sp-18E6/21, 2.5% for Sp-18E6/27, 78.0% for Ex-18E6/21, and 23.5% for Ex-18E6/27 (Table 1). Similar results were obtained for C-4I cells; at an siRNA concentration of 50 nM, survival rates were 32.5% for Sp-18E6/21, 2.0% for Sp-18E6/27, 70.5% for Ex-18E6/21, and 22.0% for Ex-18E6/27 (Table 1). Statistical analyses showed significant differences according to each type of siRNAs from the concentration of 5 nM (P<0.05). Among four types of siRNAs, the cell viability decreased with the largest scale when Sp-18E6/27 siRNA was transfected.
Sp-18E6/27 siRNA-transfected (10, 25 nM) or -untransfected HeLa cells were incubated with or without IFN-β collected from MDA-MB-468 and evaluated for the viability. In the absence of added IFN-β, the viability of HeLa cells decreased in proportion to the increase in Sp-18E6/27 siRNA concentration (52.5% at 10 nM and 21.5% at 25 nM). Also, IFN-β alone induced a rapid and dose-dependent decrease in HeLa cell viability. Notably, as shown in Fig. 1, IFN-β of low concentrations (1 and 2 mM) in combination with Sp-18E6/27 siRNAs produced the synergistic inhibitory effect to HeLa cell growth.
C-4I cells transfected with 5 nM Sp-18E6/27 siRNA and untransfected C-4I cells were cultured with different thimerosal concentrations and cell viability was determined. Compared to untreated controls (100% survival), thimerosal alone induced a dose-dependent inhibition of C-4I cell viability, decreasing the viability to 74.5% at 0.2 mM and 58.0% at 0.5 mM, respectively. As shown in Fig. 2, thimerosal decreased C-4I viability even more effectively in presence of Sp-18E6/27 siRNA, decreasing the viability to 12.5% and 4.0% at 0.2 mM and 0.5 mM, respectively.
To examine possible interactions among Sp-18E6/27 siRNA (5 nM), IFN-β (2 mM) and thimerosal (0.2 mM), we tested the effects of the three reagents in combination on the viability of HeLa and C-4I cells. Each cell type was divided into the following six experimental groups: control, 5 nM siRNAs targeting GFP; thimerosal only; thimerosal + IFN-β; siRNA + thimerosal; siRNA + IFN-β; and siRNA + thimerosal + IFN-β. In both HeLa cell and C-4I cell, the cell viability decreased significantly in group 3, 4, 5, and 6 compared to group 1 (control) (Fig. 3).
The HPV reproduce their DNAs in infected host cells through the cooperation of E6 and E7 proteins. Differences in E6 and E7 proteins between high-risk and low-risk group HPVs are mainly quantitative rather than qualitative [7]. All early HPV16 mRNAs, including those encoding E6 and E7, are transcribed by a promoter termed P97 present on the upper part of the E6 open reading frame. Eukaryocytic mRNA generally encodes genetic information for a single protein (monocistronic mRNA), however, E6 and E7 proteins are structurally generated by bicistronic E6-E7 mRNAs. With E6-E7 mRNAs, introns present in E6 open reading frames are cut to diverse sizes during the transcription process. As a result, complete E6-E7 mRNA and E6*E7 mRNA variants are formed [8]. E6 protein is formed by complete E6-E7 mRNAs, and E7 protein is formed by both complete E6-E7 mRNA and E6*E7 mRNA variants [9]. E6 and E7 proteins play different roles in the process of triggering cervical cancer, and their cooperation with each other is important in this regard. The association of E7 protein with retinoblastoma tumor-suppressor protein deactivates retinoblastoma through the inhibition of G1-S cell-cycle transition. When the abnormal proliferation is induced by E7 protein, in normal situations, the tumor suppressor protein p53 responds to cell stress by promoting cell-cycle arrest or triggering apoptosis. However, the E6 protein binds p53, and once bound, triggers ubiquitin/proteasome-mediated p53 degradation [7]. Through these actions, E6 and E7 proteins confer immortality on infected cells, thus creating opportunities for these cells to become transformed into malignant tumor cells.
Based on understanding the role of E6 and E7 proteins, researchers have made a number of attempts to genetically control the expression of these proteins as a strategy for treating HPV-related cancers. A representative method is selectively inhibiting E6/E7-encoding genes using siRNAs. The issue of applying siRNAs to cancer treatment is complicated by the fact that most tumor genes show no gross changes in their DNA sequence or minute changes, such as point mutations or translocation, during the cancer-forming process. However, the HPV genes that are a cause of cervical cancer do not have homology with human genes; thus, E6-E7 mRNAs can be specifically inhibited while minimizing adverse effects on host genes. There are numerous reports on the effects of siRNAs targeting E6-E7 mRNAs on tumor cell growth and apoptosis in HPV-positive tumors.
Jiang and Milner [10] reported that siRNA against HPV16 E6 was not sufficient to suppress cancer cell growth or compellingly inhibit apoptosis, whereas other studies reported remarkable suppression of growth and apoptosis [11]. The different outcomes of these studies likely reflects differences in the specific gene sequences targeted, methods used to inject siRNAs into nuclei, the degree of siRNA expression in nuclei, protein turnover, and the concentrations of siRNAs used. In our study, Ex-18E6 and Sp-18E6 siRNAs administered to HeLa and C-4I cell lines reduced survival rates, as described above. The inhibitory effect on survival was greater using the E6 mono-specific Sp-18E6 siRNA than using the E6-E7 specific Ex-18E6 siRNA. That is, the inhibitory effects on survival were greater in the group where only the E6 protein was selectively knocked down than in the group where both E6 and E7 proteins were inhibited. These results are consistent with the results of other studies indicating that, if E6 protein expression only is selectively inhibited and E7 protein expression is allowed to be maintained, p53 concentrations in cells can increase and induce apoptosis [12]. Initial attempts to apply dsRNAs to mammalian cells as a method for inhibiting gene expression failed owing to the fact that this strategy activated interferon pathway genes [13,14]. Later, it was found that 21-nucleotide dsRNAs inhibited the expression of mammalian cell genes without inducing interferon reactions, with 21 base-pair dsRNAs containing 3' overhangs being the most effective in vitro [15]. Subsequently, most researchers have synthesized 19-21 base pair siRNAs with 3' overhangs consisting of two bases for use in mammalian cell RNAi strategies. However, Kim et al. [16] reported that 27-nucleotide siRNAs were more effective than the commonly used 21-nucleotide siRNAs, showing that 27-nucleotide siRNAs had higher efficacy and longer durations of action. Moreover, they were effective against target areas in which 21-nucleotide siRNAs elicited no response, and they did not activate interferon or protein kinase, as was the case for 21-nucleotide siRNAs. In this study, siRNAs of two lengths-21 nucleotides (Sp-18E6/21 and Ex-18E6/21) and 27 nucleotides (Sp-18E6/27 and Ex-18E6/27)-acting on the same areas were used and tumor cell survival rates were compared. On the basis of the results obtained, we conclude that Sp-18E6/27 and Ex-18E6/27 exerted a more profound inhibition of tumor cell survival than Sp-18E6/21 and Ex-18E6/21. Taken together, our results clearly indicate that inhibitory effects on tumor cell survival were optimal when 27-nucleotide, E6 monospecific siRNAs were used.
We also addressed the effects of siRNA in conjunction with the known anti-cancer agents, IFN-b and thimerosal, on tumor cell viabilities. IFN-b, like IFN-a, is classified as a type I interferon and can effectively inhibit tumor cell growth by inducing cell differentiation and accumulation of transformed cells in S-phase, thereby effectively inducing failure to progress to G2 and M and promoting apoptosis [17]. IFN-b, administered either alone or in combination with other agents as intramuscular injections, has been used for more than 10 years to treat HPVs [18]. In a meta-analysis, Yang et al. [19] reported that, of 1,445 cases of genital warts treated with IFN-b, 27.4% to 44.4% were completely cured; there are also reports that IFN-b is effective in treating CIN-related, recurrent HPV lesions [18]. Studies on the effects of siRNAs and IFN-b in HPV infection-related cancers have not been attempted. Thimerosal (or mercurothiolate), a compound containing organic mercury, is a disinfectant for vaccines used to prevent contamination by bacteria or pathogenic microorganisms. It is widely used in various kinds of vaccine products, particularly in opened, multi-dose vaccines. Thimerosal is being studied for its potential anticancer effects in various tumors, including neuroblastomas, leukemias, and osteosarcomas [20,21]. In the current study, the inhibitory effects of IFN-b and thimerosal on tumor survival were first tested individually and these two reagents independently were effective in inhibiting the survival of HPV18-positive uterine cancer cell lines. Then, the inhibitory effects of siRNA, IFN-b and thimerosal were tested in five different combinations. The results showed that the use of all three reagents together produced the most potent inhibitory effects, demonstrating their potential for cervical cancer treatment. Importantly, combined treatment with the two reagents demonstrated a clear synergistic effect. If siRNAs can reduce the amount of drug used in anticancer chemotherapy, adverse effects associated with these chemical agents can be reduced and more effective treatment can be expected. As for the effect of RNAi on cisplatin, that is an anticancer agent commonly used in cervical cancer chemotherapy, contradictory results have been reported. Koivusalo et al. [22] suggested that the resistance of HeLa cells to cisplatin was increased by pretreatment with siRNA. In contrast, Putral et al. [23] proposed the opposite, reporting that sensitivity to cisplatin was increased by siRNA, allowing smaller doses of cisplatin to be used in cervical cancer treatment. Additional studies will be necessary to reconcile these apparent contradictions and confirm the dose-sparing effect of siRNAs.
In this study, 27-nucleotides siRNAs targeting E6-E7 mRNAs, but not E6*I-E7 mRNAs, was the most effective to inhibit the growth of HPV18-positive cervical cancer cells. Although the most effective design of siRNAs is identified, the efficient delivery of siRNAs into cells and maintenance with high concentrations are remained as unsolved problems. Nevertheless, the siRNA-based approach described in this study has the promise of making a substantial contribution to the treatment of HPV-induced cervical cancer. We suggest the necessities to find the rational design of siRNAs and effective co-factors to inhibit the growth of particular cervical cancer cells.
Figures and Tables
Fig. 1
Synergistic effect of small interfering RNA (siRNA) and interferon beta (IFN-β) to inhibit the growth of human papilloma viruse type 18-positive HeLa cells. The viability of HeLa cells decreased in proportion to the concentration of each Sp-18E6/27 siRNA and IFN-β. The combination of Sp-18E6/27 siRNAs and IFN-β produced the synergistic inhibitory effect to HeLa cell growth in low concentrations of IFN-β of 1 and 2 mM. *P<0.05.

Fig. 2
Synergistic effect of small inhibitory RNA and thimerosal to inhibit the growth of human papilloma viruse type 18-positive C-4I cells. Thimerosal alone induced a dose-dependent inhibition of C-4I cell viability. Thimerosal decreased C-4I viability even more effectively in presence of 5nM Sp-18E6/27 siRNA. *P<0.05.
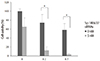
Fig. 3
Synergistic effect of small inhibitory RNA (siRNA), interferon beta (IFN-β), and thimerosal to inhibit the growth of human papilloma virus type 18-positive HeLa and C-4I cells. (A) In HeLa cell, the viability decreased the most significantly when Sp-18E6/27 siRNA (5 nM), thimerosal (0.2 mM), and IFN-β (2 mM) were combined. (B) In C-4I cell, the viability decreased the most significantly when Sp-18E6/27 siRNA (5 nM), thimerosal (0.2 mM), and IFN-β (2 mM) were combined. *P<0.05.

Table 1
The viabilities of HeLa and C-4I cells transfected with different types and concentrations of siRNAs

siRNA, small inhibitory RNA; GFP, green fluorescent protein; Sp-18E6/21, 21-nucleotide siRNA to inhibit the expression of E6 protein only; Sp-18E6/27, 27-nucleotide siRNA to inhibit the expression of E6 protein only; Ex-18E6/21, 21-nucleotide siRNA to inhibit the expression of both E6 and E7 proteins; Ex-18E6/27, 27-nucleotide siRNA to inhibit the expression of both E6 and E7 proteins.
Acknowledgments
This study was supported by a grant (2007136 and 2014559) from the Asan Institute for Life Sciences, Seoul, Korea.
References
1. Bosch FX, de Sanjose S. The epidemiology of human papillomavirus infection and cervical cancer. Dis Markers. 2007; 23:213–227.
2. Manavi M, Hudelist G, Fink-Retter A, Gschwantler-Kaulich D, Pischinger K, Czerwenka K. Human papillomavirus DNA integration and messenger RNA transcription in cervical low- and high-risk squamous intraepithelial lesions in Austrian women. Int J Gynecol Cancer. 2008; 18:285–294.
3. Khan MJ, Castle PE, Lorincz AT, Wacholder S, Sherman M, Scott DR, et al. The elevated 10-year risk of cervical precancer and cancer in women with human papillomavirus (HPV) type 16 or 18 and the possible utility of type-specific HPV testing in clinical practice. J Natl Cancer Inst. 2005; 97:1072–1079.
4. Munger K, Baldwin A, Edwards KM, Hayakawa H, Nguyen CL, Owens M, et al. Mechanisms of human papillomavirus-induced oncogenesis. J Virol. 2004; 78:11451–11460.
5. Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001; 411:494–498.
6. Amarzguioui M, Holen T, Babaie E, Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003; 31:589–595.
7. Ghittoni R, Accardi R, Hasan U, Gheit T, Sylla B, Tommasino M. The biological properties of E6 and E7 oncoproteins from human papillomaviruses. Virus Genes. 2010; 40:1–13.
8. Magaldi TG, Almstead LL, Bellone S, Prevatt EG, Santin AD, DiMaio D. Primary human cervical carcinoma cells require human papillomavirus E6 and E7 expression for ongoing proliferation. Virology. 2012; 422:114–124.
9. Grassmann K, Rapp B, Maschek H, Petry KU, Iftner T. Identification of a differentiation-inducible promoter in the E7 open reading frame of human papillomavirus type 16 (HPV-16) in raft cultures of a new cell line containing high copy numbers of episomal HPV-16 DNA. J Virol. 1996; 70:2339–2349.
10. Jiang M, Milner J. Selective silencing of viral gene expression in HPV-positive human cervical carcinoma cells treated with siRNA, a primer of RNA interference. Oncogene. 2002; 21:6041–6048.
11. Niu XY, Peng ZL, Duan WQ, Wang H, Wang P. Inhibition of HPV 16 E6 oncogene expression by RNA interference in vitro and in vivo. Int J Gynecol Cancer. 2006; 16:743–751.
12. DeFilippis RA, Goodwin EC, Wu L, DiMaio D. Endogenous human papillomavirus E6 and E7 proteins differentially regulate proliferation, senescence, and apoptosis in HeLa cervical carcinoma cells. J Virol. 2003; 77:1551–1563.
13. Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998; 67:227–264.
14. Minks MA, Benvin S, Maroney PA, Baglioni C. Synthesis of 2'5'-oligo(A) in extracts of interferon-treated HeLa cells. J Biol Chem. 1979; 254:5058–5064.
15. Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001; 20:6877–6888.
16. Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005; 23:222–226.
17. Chiantore MV, Vannucchi S, Accardi R, Tommasino M, Percario ZA, Vaccari G, et al. Interferon-β induces cellular senescence in cutaneous human papilloma virus-transformed human keratinocytes by affecting p53 transactivating activity. PLoS One. 2012; 7:e36909.
18. Gonzalez-Sanchez JL, Martinez-Chequer JC, Hernandez-Celaya ME, Barahona-Bustillos E, Andrade-Manzano AF. Randomized placebo-controlled evaluation of intramuscular interferon beta treatment of recurrent human papillomavirus. Obstet Gynecol. 2001; 97:621–624.
19. Yang J, Pu YG, Zeng ZM, Yu ZJ, Huang N, Deng QW. Interferon for the treatment of genital warts: a systematic review. BMC Infect Dis. 2009; 9:156.
20. Parran DK, Barker A, Ehrich M. Effects of thimerosal on NGF signal transduction and cell death in neuroblastoma cells. Toxicol Sci. 2005; 86:132–140.
21. Woo KJ, Lee TJ, Bae JH, Jang BC, Song DK, Cho JW, et al. Thimerosal induces apoptosis and G2/M phase arrest in human leukemia cells. Mol Carcinog. 2006; 45:657–666.
22. Koivusalo R, Krausz E, Helenius H, Hietanen S. Chemotherapy compounds in cervical cancer cells primed by reconstitution of p53 function after short interfering RNAmediated degradation of human papillomavirus 18 E6 mRNA: opposite effect of siRNA in combination with different drugs. Mol Pharmacol. 2005; 68:372–382.
23. Putral LN, Bywater MJ, Gu W, Saunders NA, Gabrielli BG, Leggatt GR, et al. RNA interference against human papillomavirus oncogenes in cervical cancer cells results in increased sensitivity to cisplatin. Mol Pharmacol. 2005; 68:1311–1319.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download