Abstract
Ossification of the posterior longitudinal ligament of the spine (OPLL) is a common disease in aging populations and sometimes results in serious neurological problems due to compression of the spinal cord and nerve roots. OPLL is a multi-factorial (polygenic) disease controlled by genetic and environmental factors. Studies searching for the genetic component of OPLL, using linkage and association analyses, are in progress and several susceptibility genes have been reported. This paper reviews the recent progress in the genetic study of OPLL and comments on its future task.
The posterior longitudinal ligament of the spine (PLL) is a ligament that runs behind the spinal column (vertebral bodies and intervertebral discs). PLL is situated anterior to the spinal cord within the spinal canal. Ossification of the PLL (OPLL; MIM 602475) is a disease state caused by ectopic ossification. OPLL is a common disease. The incidence of OPLL is 1.9-4.3% in Japan.[1,2] Comparable incidence has been reported in other countries, especially in East Asia.[3] The average age of onset is over 50 years with male predominance.[4] OPLL presents with neurological symptoms due to compression of spinal cord and nerve roots as well as neuropathic pain and stiffness of the neck and trunk. These symptoms affect motility and quality of life of the patients.
From the etiological point of view, OPLL is divided into 2 categories; primary (idiopathic) and secondary (syndromic). The latter includes OPLL associated with monogenic diseases like hypophosphatemic rickets/osteomalacia. Several forms of hypophosphatemic rickets are known, including an X-linked form (MIM 307800) caused by phosphate regulating endopeptidase homolog, X-linked (PHEX) mutations (MIM 300550), an autosomal dominant form (MIM 193100) caused by fibroblast growth factor 23 (FGF23) mutations (MIM 605380), an X-linked recessive form (MIM 300554) caused by chloride channel, voltage-sensitive 5 (CLCN5) mutations (MIM 300008), and autosomal recessive forms caused by dentin matrix acidic phosphoprotein 1 (DMP1) (MIM 600980), hypophosphatemic rickets, autosomal recessive 2 (ARHR2) (MIM 613312) or ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1) (MIM 173335) mutations. 'tiptoe walking' (TTW) mouse, which has a spontaneous nonsense mutation in ENPP1 is a good model for OPLL.[5] Also, OPPL is a frequent complication in patients with endocrine disorders including hypoparathyroidism[6] and acromegaly/gigantism.[7] However, most cases of OPLL are idiopathic. Therefore, I refer to idiopathic OPLL hereafter.
Many reports on the underlying mechanisms of OPLL have suggested that OPLL is a multi-factorial (polygenic) disease influenced by genetic and environmental (non-genetic) factors. Several clinical factors including age,[8] diabetes mellitus (DM)[9] and obesity[10] have been reported as risk factors for OPLL. In addition, vitamin A-rich diet, exercise and abnormal mechanical stress to the head have been considered as environmental factors for OPLL.[7] On the other hand, OPLL has a strong genetic preposition. A study using 347 OPLL families reported a prevalence of OPPL of 26% in the parents of the probands and 29% in the sibs.[11] Matsunaga et al.[12] studied the association between OPLL and human leukocyte antigen (HLA) haplotypes in families of 24 patients with OPLL and found higher prevalence of OPLL in the siblings showing a higher share of identical HLA haplotypes. As in other multi-factorial diseases, genome studies are revealing the genetic factors of OPLL. A lot of linkage and association studies have been conducted and many genes/loci that link to OPLL susceptibility have been reported (Table 1).
The first one was a sib-pair linkage analysis conducted by a Utah group,[13] which examined 53 families by a non-parametric linkage analysis focusing on the HLA region and found a significant linkage on D6S276 (P=6×10-6). Subsequently, by a candidate gene approach using in 280 patients and 210 controls for positional candidates around the marker, they found an association with collagen, type XI, alpha 2 (COL11A2) (P=4×10-4). COL11A2 (MIM 120290) encodes one of the 3 α-chains of type XI collagen, a cartilage-specific collagen. The group also reported association (P=0.0028) with retinoid X receptor, beta (RXRB) (MIM 180246) adjacent to COL11A2.[14]
A group led by Inoue expanded on the study by increasing the number of sibs and found a significant linkage at D21S1903 on 21q by a genome-wide linkage study.[15] They conducted an association study of 150 candidate genes in a 20-Mb region around the marker using 280 OPLL patients and 210 controls, and found association with collagen, type VI, alpha 1 (COL6A1) (P=3×10-6). COL6A1 (MIM 120290) encodes one of the 3 α-chains of type VI collagen. Furushima et al.[16] performed a linkage study for candidate genes selected from expression profiles during osteoblastic differentiation of human mesenchymal stem cells and found suggestive evidence of linkage with bone morphogenetic protein 4 (BMP4) (MIM 112262).
Those studies are interesting but were dependent on small number of samples (172 at the most), and most of the subjects were collected in very limited areas. Karasugi et al.[17] performed a large-scale genome-wide linkage study using 410 Japanese OPLL individuals (214 affected sib-pairs); however, they could not replicate the previous linkage results nor find any new loci. In stratification analyses for definite cervical OPLL that included subjects with more than 2 ossified vertebrae only, they found loci with suggestive linkage on 1p, 2p, 7q, 16q, and 20p. Fine mapping using additional markers detected the highest non-parametric lod (NPL) score (3.43, P=0.00027) at D20S894 on chromosome 20p12 in a subgroup that had no complication of DM.
Several groups worked on candidate gene association studies. A number of genes/loci associated with the OPLL susceptibility have been reported, including genes for nucleotide pyrophosphatase/phosphodiesterases (NPPS)/ENNP1[18], transforming growth factor (TGF)-β1[19], estrogen receptor (ESR),[20] interleukin 1, beta (IL-1β),[20] vitamin D receptor (VDR),[21] bone morphogenetic protein 2 (BMP2),[22] runt-related transcription factor 2 (RUNX2),[23] toll-like receptor 5 (TLR5),[24] interleukin 15 receptor, alpha (IL-15RA),[25] and BMP9 [26] (Table 1). However, the results of these studies are not sufficiently convincing because of their small sample sizes, small number of sequence variants examined and lack of functional proof of the variants and/or genes. Few variants per gene (usually only one single nucleotide polymorphism [SNP]) were examined; the statistical significance of their association is not sufficient judging by current standards.
At present, the largest study is the case-control association study that examined 109 sequence polymorphisms in 35 candidate genes using a ~1,600 case-control cohort and found the association of TGF beta 3 (TGFB3) (P=0.00040).[27] TGFB3 (MIM190230) is a well-known gene related to osteogenesis and located in the weak linkage region identified by the previous linkage study;[15] however, the association has not been replicated in other studies to my knowledge. Like other susceptibility genes so far reported, replication studies with decent scale are necessary for the association.
The results of Karasugi et al.[17] indicate that OPLL is genetically heterogeneous, which is consistent with the vast diversity of its clinical features, including sex predominance, age at onset and prognosis by location of the lesion (i.e., cervical, thoracic, lumbar) and type of ossification (i.e., continuous, segmental, mixed). By stratification, i.e., subgroup analysis based on clinical and demographic parameters, we can reduce the heterogeneity of the cases and hence expect to increase the power of detection in association studies. However, stratification is a trade-off with a decrease of the sample number. Larger scale studies enrolling thousands of subjects will be necessary. As linkage studies have a theoretical limitation in pinpointing the location of the susceptibility gene, association studies with high-density SNPs should be the future strategy. Like in other common bone and joint diseases,[28,29,30,31] genome-wide association study (GWAS) is awaited. Whole exome and whole genome sequencing are also promising approaches.
Since OPLL is a multi-factorial disease, both genetic and environmental factors must be clarified for better understanding of its etiology and pathology as well as for correct diagnosis, prediction of prognosis and effective treatment of the patients. One of the important future tasks is a longitudinal study of cohorts with detailed clinical information that could evaluate environmental factors based on the adjustment of genetic factors by genotyping results. In this point, larger scale studies will also be necessary. To accomplish such tasks within a certain period of time, international collaboration is the only way to go. I am optimistic because international collaborations have succeeded in many association studies of bone and joint diseases.[32,33,34,35]
Figures and Tables
Table 1
Previously reported ossification of the posterior longitudinal ligament of the spine susceptibility genes
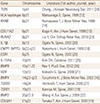
TLR, toll-like receptor; RXRB, retinoic X receptor β; COL, collagen; RUNX, runt-related transcription factor; IL, interleukin; ENPP, ectonucleotide pyrophosphatase/phosphodiesterase; NPPS, nucleotide pyrophosphatase; ESR, estrogen receptor; VDR, vitamin D (1,25-dihydroxyvitamin D3) receptor; BMP, bone morphogenetic protein; TGFB, transforming growth factor-beta.
Notes
References
1. Matsunaga S, Sakou T. Overview of epidemiology and genetics. In : Yonenobu K, Nakamura K, Toyama Y, editors. Ossification of the posterior longitudinal ligament. 2nd ed. Tokyo: Springer;2006. p. 7–9.
2. Matsunaga S, Sakou T. Ossification of the posterior longitudinal ligament of the cervical spine: etiology and natural history. Spine (Phila Pa 1976). 2012; 37:E309–E314.
3. Saetia K, Cho D, Lee S, et al. Ossification of the posterior longitudinal ligament: a review. Neurosurg Focus. 2011; 30:E1.

4. Ohtsuka K, Terayama K, Yanagihara M, et al. A radiological population study on the ossification of the posterior longitudinal ligament in the spine. Arch Orthop Trauma Surg. 1987; 106:89–93.

5. Okawa A, Nakamura I, Goto S, et al. Mutation in Npps in a mouse model of ossification of the posterior longitudinal ligament of the spine. Nat Genet. 1998; 19:271–273.

6. Okazaki T, Takuwa Y, Yamamoto M, et al. Ossification of the paravertebral ligaments: a frequent complication of hypoparathyroidism. Metabolism. 1984; 33:710–713.

7. Taguchi T. Etiology and pathogenesis. In : Yonenobu K, Nakamura K, Toyama Y, editors. Ossification of the posterior longitudinal ligament. 2nd ed. Tokyo: Springer;2006. p. 33–35.
8. Wu JC, Liu L, Chen YC, et al. Ossification of the posterior longitudinal ligament in the cervical spine: an 11-year comprehensive national epidemiology study. Neurosurg Focus. 2011; 30:E5.

9. Kobashi G, Washio M, Okamoto K, et al. High body mass index after age 20 and diabetes mellitus are independent risk factors for ossification of the posterior longitudinal ligament of the spine in Japanese subjects: a case-control study in multiple hospitals. Spine (Phila Pa 1976). 2004; 29:1006–1010.

10. Shingyouchi Y, Nagahama A, Niida M. Ligamentous ossification of the cervical spine in the late middle-aged Japanese men. Its relation to body mass index and glucose metabolism. Spine (Phila Pa 1976). 1996; 21:2474–2478.

11. Terayama K. Genetic studies on ossification of the posterior longitudinal ligament of the spine. Spine (Phila Pa 1976). 1989; 14:1184–1191.

12. Matsunaga S, Yamaguchi M, Hayashi K, et al. Genetic analysis of ossification of the posterior longitudinal ligament. Spine (Phila Pa 1976). 1999; 24:937–939.

13. Koga H, Sakou T, Taketomi E, et al. Genetic mapping of ossification of the posterior longitudinal ligament of the spine. Am J Hum Genet. 1998; 62:1460–1467.

14. Numasawa T, Koga H, Ueyama K, et al. Human retinoic X receptor beta: complete genomic sequence and mutation search for ossification of posterior longitudinal ligament of the spine. J Bone Miner Res. 1999; 14:500–508.

15. Tanaka T, Ikari K, Furushima K, et al. Genomewide linkage and linkage disequilibrium analyses identify COL6A1, on chromosome 21, as the locus for ossification of the posterior longitudinal ligament of the spine. Am J Hum Genet. 2003; 73:812–822.

16. Furushima K, Shimo-Onoda K, Maeda S, et al. Large-scale screening for candidate genes of ossification of the posterior longitudinal ligament of the spine. J Bone Miner Res. 2002; 17:128–137.

17. Karasugi T, Nakajima M, Ikari K, et al. A genome-wide sib-pair linkage analysis of ossification of the posterior longitudinal ligament of the spine. J Bone Miner Metab. 2013; 31:136–143.

18. Nakamura I, Ikegawa S, Okawa A, et al. Association of the human NPPS gene with ossification of the posterior longitudinal ligament of the spine (OPLL). Hum Genet. 1999; 104:492–497.

19. Kamiya M, Harada A, Mizuno M, et al. Association between a polymorphism of the transforming growth factor-beta1 gene and genetic susceptibility to ossification of the posterior longitudinal ligament in Japanese patients. Spine (Phila Pa 1976). 2001; 26:1264–1267.
20. Ogata N, Koshizuka Y, Miura T, et al. Association of bone metabolism regulatory factor gene polymorphisms with susceptibility to ossification of the posterior longitudinal ligament of the spine and its severity. Spine (Phila Pa 1976). 2002; 27:1765–1771.

21. Kobashi G, Ohta K, Washio M, et al. FokI variant of vitamin D receptor gene and factors related to atherosclerosis associated with ossification of the posterior longitudinal ligament of the spine: a multi-hospital case-control study. Spine (Phila Pa 1976). 2008; 33:E553–E558.
22. Wang H, Liu D, Yang Z, et al. Association of bone morphogenetic protein-2 gene polymorphisms with susceptibility to ossification of the posterior longitudinal ligament of the spine and its severity in Chinese patients. Eur Spine J. 2008; 17:956–964.

23. Liu Y, Zhao Y, Chen Y, et al. RUNX2 polymorphisms associated with OPLL and OLF in the Han population. Clin Orthop Relat Res. 2010; 468:3333–3341.

24. Chung WS, Nam DH, Jo DJ, et al. Association of toll-like receptor 5 gene polymorphism with susceptibility to ossification of the posterior longitudinal ligament of the spine in korean population. J Korean Neurosurg Soc. 2011; 49:8–12.

25. Kim DH, Jeong YS, Chon J, et al. Association between interleukin 15 receptor, alpha (IL15RA) polymorphism and Korean patients with ossification of the posterior longitudinal ligament. Cytokine. 2011; 55:343–346.

26. Ren Y, Liu ZZ, Feng J, et al. Association of a BMP9 haplotype with ossification of the posterior longitudinal ligament (OPLL) in a Chinese population. PLoS One. 2012; 7:e40587.

27. Horikoshi T, Maeda K, Kawaguchi Y, et al. A large-scale genetic association study of ossification of the posterior longitudinal ligament of the spine. Hum Genet. 2006; 119:611–616.

28. Nakajima M, Takahashi A, Kou I, et al. New sequence variants in HLA class II/III region associated with susceptibility to knee osteoarthritis identified by genome-wide association study. PLoS One. 2010; 5:e9723.

29. Kou I, Takahashi A, Urano T, et al. Common variants in a novel gene, FONG on chromosome 2q33.1 confer risk of osteoporosis in Japanese. PLoS One. 2011; 6:e19641.

30. Takahashi Y, Kou I, Takahashi A, et al. A genome-wide association study identifies common variants near LBX1 associated with adolescent idiopathic scoliosis. Nat Genet. 2011; 43:1237–1240.

31. Song YQ, Karasugi T, Cheung KM, et al. Lumbar disc degeneration is linked to a carbohydrate sulfotransferase 3 variant. J Clin Invest. 2013; 123:4909–4917.

32. Miyamoto Y, Mabuchi A, Shi D, et al. A functional polymorphism in the 5' UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat Genet. 2007; 39:529–533.

33. Hwang JY, Lee SH, Go MJ, et al. Meta-analysis identifies a MECOM gene as a novel predisposing factor of osteoporotic fracture. J Med Genet. 2013; 50:212–219.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download