Abstract
Backgrounds/Aims
Cysteine dioxygenase type 1 (CDO1) acts as a tumor suppressor and is silenced by promoter methylation in various malignancies. The relationship between the CDO1 methylation status and hepatocellular carcinoma (HCC) tumorigenesis was evaluated.
Methods
Using a HCC cell line (SNU423), an in vitro demethylation study was performed to confirm whether promoter methylation causes CDO1 down-regulation. The SNU423 cells transfected with the CDO1 cell function was compared to that of naïve cells. An in vivo study using immunohistochemical staining of HCC specimens that were collected from patients who underwent curative liver resection was also performed.
Results
CDO1 was activated after demethylation treatment in the HCC specimens. Moreover, tumor cell proliferation, colony-forming, migration, and invasion activities significantly decreased after CDO1 transfection (p<0.05). The percentage of tumors that were larger than 5 cm was higher in patients who had a lower expression of CDO1 (p=0.030). Vascular invasion and histological grade were independent prognostic factors for poor overall and recurrence-free survival. The degree of CDO1 expression was not an independent prognostic factor in this study's population.
Hepatocellular carcinoma (HCC) is a common malignancy and the fourth most common cause of cancer-related death in South Korea. HCC is difficult to treat and associated with poor prognoses and high mortality rates due to frequent recurrence. Many clinical factors, such as the advanced pathological tumor-node-metastasis (pTNM) stage and liver function status, are associated with the post-treatment prognosis.1 In addition, many recent studies have demonstrated that there is a strong correlation between the cancer prognosis and genetic factors.
The term ‘epigenetics’ is used now to describe the study of stable alterations in gene expression potential that arise during cell development and proliferation. Epigenetic changes can induce an oncotic occurrence and development by altering the levels of gene expressions, even in the absence of variations in the DNA sequences of specific genes.2 DNA methylation is the most common type of ‘epigenetic change’ and is the subject of much research. Moreover, DNA methylation of tumor suppressor genes can be used as a biomarker to improve the clinical diagnosis or prognosis prediction in patients with cancer.345
Cystein dioxygenase (CDO) is a non-heme structured, iron-containing metalloenzyme involved in the conversion of toxic cysteine to cysteine sulfate, and in taurine biosynthesis in mammals.6 There are two types of CDO enzymes. Type I CDO (CDO1) is present in the cytosol; whereas, type II CDO is found in the cell membrane. The gene encoding CDO1 is located on chromosome 5 q23.2, and many studies have shown that deletion or epigenetic silencing of this chromosomal region contributes to tumorigenesis. CDO1 has also been shown to regulate protein function and antioxidant defense mechanisms through the oxidation of cysteine7: CDO1 expression is elevated in the liver and placenta, but decreased in the heart, brain, and pancreas.
Recently, many studies have demonstrated that CDO1 functions as a tumor suppressor and is silenced by promoter methylation in many malignancies.8 For example, Dietrich et al. found that CDO1 promoter methylation is a biomarker for prognosis in patients with anthracycline-treated, estrogen receptor-positive, lymph node-positive breast cancer.9 Additionally, Jeschke et al. reported that inactivation of CDO1 frequently contributes to the survival of breast cancer cells and resistance to anthracyclines.10 Yang et al.11 evaluated whether the methylation status of the CDO1 promoter was of diagnostic value for hepatitis B virus-related HCC. However, the methylation status of CDO1 in HCC remains unknown.
Using a microarray analysis of the HCC tissues from patients who underwent curative liver resection, our research team identified several genes that are abnormally expressed in HCC. These results identified CDO1 as a gene that is suppressed in the HCC tissues compared to normal liver cells.12
Therefore, the aim of this study was to identify correlations between CDO1 methylation and HCC, using HCC cell lines, and to confirm whether CDO1 methylation status is relevant to the prognosis of patients with HCC.
Human HCC cell lines, SNU387, SNU423, SNU449, SNU475, and SK-Hep-1, were purchased from the Korean Cell Line Bank (KCLB) in Korea. The cells were cultured in RPMI1640 and the DMEM medium was supplemented with 10% (v/v) FBS and antibiotics (100 units/ml penicillin and 100 µg/ml streptomycin) and grown at 37℃ under a 5% CO2 atmosphere.
RNA was extracted from all cell lines using a RNeasy mini kit (Qiagen, Cambridge, MA) according to the manufacturer's protocol. The cDNA was produced by the reverse transcription of 2 µg total RNA using an oligo-dT primer and superscript II reverse transcriptase (Invitrogen, Carlsbad, CA). The RT-PCR experiments were carried out with the following primers: CDO1 R-AGTGAAGGCTCACAGCAGGT and F-TCTCTGTTGGGGTGAAGGAC. We used GAPDH-specific primers as an internal control: R-TGCTGTAGCCAAATTCGTTG and F-GTCAGTGGTGGACCTGACCT.
The human HCC cell lines that exhibited no or weak expression (SNU387, SNU423, SNU449, SNU475, and SK-Hep1) of CDO1 were treated with the demethylation agent 70 µM 5-aza-2′-deoxycytidine (5-Aza-dC, Sigma, St. Louis, MO), and grown for 4 days. The gene expression and DNA methylation patterns before and after treatment with the demethylating agent were examined by the RT-PCR. After the demethylation test, we selected the SNU423 cell line for further in vitro studies.
A plasmid vector that expressed the full-length human CDO1 (pcDNA3-CDO1) was constructed by transfecting CDO1 cDNA into a pcDNA3 expression vector (Invitrogen, Carlsbad, CA). All plasmid sequences were confirmed by sequencing analysis. The transfection of plasmids was performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA).
For the western blot analysis, cells were washed with cold PBS, and the collected cells were extracted in a RIPA buffer (Thermo Fisher Scientific, Rockford, IL) and quantified using the Bicinchoninic acid (BCA) method. The protein samples (40 µg) were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). After transfer, the nitrocellulose (NC) membranes were incubated with a primary antibody (for CDO1; Abcam, Cambridge, MA) and secondary antibody (Santa Cruz Biotech, CA) and visualized using enhanced chemiluminescence (ECL) (Thermo Fisher Scientific, Rockford, IL). The relative protein levels were calculated using β-actin as a loading control.
Cell growth was determined by MTS assay by using a CellTiter 96®AQueous One Solution Cell Proliferation Assay Kit (G3580, Promega). In brief, 2×103 cells (per well) were seeded in 96-well plates after transfection. After 1, 3, 5, and 7 days, 10 µl of the MTS solution was added to each well. Plates were incubated for an additional 1 hour at 37℃, after which the absorbance at 490 nm was recorded using a microplate reader (BioTek, Winooski, VT) to calculate the cell survival percentages. For the colony formation assay, cells were plated at 1×103 cells/well in 6-well plates, incubated for 2 weeks in a complete growth medium, and stained with 0.2% crystal violet. The colonies were photographed and counted after 2 weeks of incubation at 37℃. All assays were performed in triplicates.
Migration assays were performed using a 35-mm µ-dish (ibidi, Martinsried, Germany) according to the manufacturer's protocol. Briefly, the 5×105 cells were placed into two-chamber cell culture inserts in µ-dishes. After cell attachment, the culture inserts were gently removed and cell migration was evaluated by light microscopy at the indicted time intervals. All assays were performed in triplicates.
The cell invasion assay was performed using a 24-well Transwell system (8-µm, BD Biosciences, Franklin Lakes, NJ). The cells were starved in a serum-free medium overnight, trypsinized, and washed three times in RPMI1640 without FBS. The cells (2×105) were seeded into the upper chamber, and 600 µl of RPMI1640 containing 10% FBS was placed in the lower chamber. After 48 hours of incubation, the cells remaining in the upper chamber were removed. The cells on the lower surface of the membrane were fixed in 4% paraformaldehyde and stained with 0.2% crystal violet. Stained cells were photographed. All assays were performed in triplicates.
HCC specimens were collected from 182 patients who underwent curative liver resection at the Korea Cancer Center Hospital from January 1997 to May 2004. Patients provided informed consent for the use of their tissues, and the study was approved by the Institutional Review Board (IRB) of the Korea Institute of Radiological and Medical Sciences (KIRAMS). The degree of CDO1 protein expression in HCC tissues was analyzed using immunohistochemical staining.
IH staining was performed using a tissue microarray technique. CDO1 protein expression was detected in sections of the paraffin-embedded HCC tissues using a CDO1 primary antibody. The immunostained slides of the HCC tissues were graded by a pathologist on a scale of 0 to 3 according to the intensity of the CDO1 protein staining, which ranged from 0% to 100%. The immunohistochemistry (IH) index was calculated based on the immunostaining intensity and proportion of stained cells (intensity×% population, range 0–300). Statistical analyses of the relationships between patient clinical variables, including the IH index, and survival rates were conducted. An IH index of 0 indicated no CDO1 protein expression; whereas, an IH index of 300 indicated that the amount of CDO1 protein expression was equal to that in the normal liver tissue cells.
We analyzed all data using the Statistical Package for the Social Sciences (SPSS) with the Standard for Medical Services (Medical Plus Pack) version 23 software. The IH index and clinical variables, including the Edmonson-Steiner histological grade, tumor number, tumor size, preoperative alpha-fetoprotein, microvessel invasion, and gross vascular invasion, were compared using chi-square tests. Differences with p-values of less than 0.05 were considered statistically significant.
CDO1 expression was enhanced in all of the HCC cell lines following demethylation treatment. However, the degree of gene expression varied (Fig. 1). Thus, the downregulation of CDO1 in the HCC cells was due to epigenetic methylation. Among the HCC cell lines tested, the SNU423 cell line was selected for further experiments.
To study the tumor-suppressive capacity of CDO1, CDO1 was transfected into a CDO1-suppressed HCC cell line (SNU423). CDO1 overexpression was confirmed by western blotting (Fig. 2A). A subsequent analysis revealed that the proliferation, colony-forming, migration, and invasion activities of the HCC cells were suppressed by CDO1 overexpression compared with the control cells (Figs. 2B, 2C, 2D, and 2E).
The CDO1 protein staining intensity varied greatly between the HCC tissues. The representative images of the various staining intensities are displayed in Fig. 3. A dot plot of staining intensity and the proportions of stained cells in each HCC tissue was generated (Fig. 4). The number of patients with a staining intensity with a grade of 0–1 was 121 (66.5%), and the number of patients with less than 50% of stained cells was 104 (57.1%). Thus, CDO1 protein expression was suppressed in the majority of the HCC tissues (Fig. 4). Based on this analysis, patients were divided into two groups according to IH index value; 150 patients had an IH index of 100 or less; whereas 32 patients exhibited an IH index of greater than 100. Notably, an IH index of 100 or less was significantly associated with an HCC size of greater than 5 cm (47.3% versus 28.1%, respectively; p=0.030) (Table 1). The independent risk factors that affected overall survival were microvascular (p<0.001) and macrovascular invasions (p=0.020). In addition, the independent risk factors that affected HCC recurrence were microvascular invasion (p<0.001), macrovascular invasion (p=0.023), histologic grade (p=0.005), and a tumor size of more than 5 cm (p=0.035). However, the degree of CDO1 protein expression was not an independent risk factor for overall or recurrence-free survival.
DNA methylation is a key mechanism that inhibits the expression of tumor-suppressor genes and the most frequently studied epigenetic alteration.3 CG-rich regions, known as cytosine-phosphate-guanine (CpG) islands, are located within 5′ of the ends of genes, which includes the promoter region, untranslated region, and first exon, and can affect the activity of tumor-suppressor genes.13 The CpG islands are not usually methylated in healthy cells13; however, aberrant hypermethylation of the CpG islands, which leads to transcriptional inactivation and gene silencing, is a frequent early event in carcinogenesis and considered a common mechanism of the loss of the tumor-suppressor gene function in human cancers.314 Epigenetic alterations, such as DNA methylation, are thought to predispose an individual to genetic alterations during tumorigenesis. Furthermore, the hypermethylation of tumor-suppressor genes is associated with tumor cell proliferation, migration, invasion, and colony formation.15 For example, the promoter CpG island methylation of CDO1 has been found in multiple tumor types, including breast, esophagus, lung, bladder, and stomach cancer8; however, to date, no reports have described studies of CDO1 methylation in HCC tissues and cell lines.
The development and progression of HCC is a multi-step process that involves a sequence that ranges from chronic hepatitis to cirrhosis to HCC.16 Moreover, the aberrant methylation of tumor-suppressor genes may play an important role in the course of this multi-step hepatocarcinogenesis.17 Hu et al.18 reported that reduced phosphatase and tensin homolog (PTEN) expression levels are involved in the pathogenesis of HCC and that decreased PTEN expression is correlated with tumor progression, high alpha-fetoprotein levels, p53 overexpression, and poor prognosis in patients with HCC. Nishida et al.17 reported an analysis of multiple tumor-suppressor genes, including HIC-1, CDH1, and MINT1, and Um et al.19 also reported a study of multiple genes, including APC, SOCS1, and COX2, and concluded that a general stepwise increase in methylation events occurred during a hepatitis B virus-related multistep hepatocarcinogenesis. Additionally, epigenetic changes may occur predominantly in the early stages of HCC development.19
In this study, CDO1 expression was suppressed in the HCC cell lines and was increased after treatment with the demethylation agent, 5-Aza-dC. Thus, the suppression of CDO1 expression occurred through promoter methylation. We confirmed that the overexpression of CDO1 strongly inhibited cell proliferation, colony-forming, migration, and invasion activities in the HCC cell lines. We also demonstrated that CDO1 expression was suppressed in the HCC tissues that were collected from patients who had undergone liver resection. These findings implied that promoter methylation may lead to the suppression of CDO1 expression in HCC tissues.
Indeed, this study demonstrated that CDO1 expression was suppressed by promoter methylation in patients with HCC. Further analysis displayed that CDO1 suppression was related to tumor size, with greater suppression of CDO1 expression in patients with HCC tumors of more than 5 cm in size; these results were supported by the IH index analysis. This result may imply that the suppression of CDO1 expression by promoter methylation was inversely related to the growth of HCC. Taken together, these findings supported that CDO1 acts as a tumor-suppressor gene in HCC.
Notably, the degree of CDO1 expression was not related to patient prognosis. Many factors, such as microvessel invasion, satellite nodules, large tumor size, advanced pTNM stage, active hepatitis activity, peri-operative transfusion, and tumor biologic factors (proliferative and angiogenic activities), are known to influence the prognosis in patients with HCC after liver resection.20 Additionally, epigenetic changes such as DNA methylation may occur predominantly in the early stages of HCC development.20 Therefore, CDO1 expression has a high probability of influencing the early steps of tumorigenesis in HCC, and we inferred that the degree of CDO1 expression was not an independent factor for HCC patient prognosis.
In conclusion, our results suggested that methylation down-regulated the expression of CDO1 in HCC cells and that CDO1 methylation may be a valuable diagnostic biomarker for HCC. However, the key regulatory mechanisms through which CDO1 may exert its tumor-suppressive functions are still unknown. Therefore, further studies are needed to assess these mechanisms.
Figures and Tables
 | Fig. 1CDO1 expression. CDO1 expression was evaluated in the HCC cell lines with and without demethylation (5-Aza-dC) treatment by RT-PCR. (GAPDH, glyceraldehyde 3-phospate dehydrogenase: housekeeping gene). |
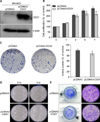 | Fig. 2Analyses of CDO1 protein in the SNU423 HCC cell line with and without pCDNA3-CDO1 transfection (CDO1 overexpression). (A) Western blot analysis of CDO1 protein in SNU423 HCC cells; (B) Cell proliferation assay: The proliferation activities of HCC cells with pCDNA3-CDO1 transfection were suppressed at day 5 (p<0.025) and at day 7 (p<0.003); (C) Colony-forming assay: The colony-forming activities of the HCC cells with pCDNA3-CDO1 transfection were suppressed after 2 weeks (p<0.014); (D) migration assay; (E) invasion assay. |
 | Fig. 3Immunohistochemical analysis for the detection of the CDO1 protein in HCC tissues. Representative samples of the different staining intensities are displayed. |
ACKNOWLEDGEMENTS
This study was supported by a grant from the Korea Institute of Radiological and Medical Sciences (KIRAMS), which was funded by the Ministry of Science, Information, and Communications Technique (ICT) and Future Planning, Republic of Korea (grant no. 1711031800).
References
1. Belghiti J, Regimbeau JM, Durand F, Kianmanesh AR, Dondero F, Terris B, et al. Resection of hepatocellular carcinoma: a European experience on 328 cases. Hepatogastroenterology. 2002; 49:41–46.
2. Wolffe AP, Matzke MA. Epigenetics: regulation through repression. Science. 1999; 286:481–486.
3. Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002; 3:415–428.
4. Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000; 16:168–174.
5. Paz MF, Fraga MF, Avila S, Guo M, Pollan M, Herman JG, et al. A systematic profile of DNA methylation in human cancer cell lines. Cancer Res. 2003; 63:1114–1121.
6. Tsuboyama-Kasaoka N, Hosokawa Y, Kodama H, Matsumoto A, Oka J, Totani M. Human cysteine dioxygenase gene: structural organization, tissue-specific expression and downregulation by phorbol 12-myristate 13-acetate. Biosci Biotechnol Biochem. 1999; 63:1017–1024.
7. Oien DB, Moskovitz J. Ablation of the mammalian methionine sulfoxide reductase A affects the expression level of cysteine deoxygenase. Biochem Biophys Res Commun. 2007; 352:556–559.
8. Brait M, Ling S, Nagpal JK, Chang X, Park HL, Lee J, et al. Cysteine dioxygenase 1 is a tumor suppressor gene silenced by promoter methylation in multiple human cancers. PLoS One. 2012; 7:e44951.
9. Dietrich D, Krispin M, Dietrich J, Fassbender A, Lewin J, Harbeck N, et al. CDO1 promoter methylation is a biomarker for outcome prediction of anthracycline treated, estrogen receptor-positive, lymph node-positive breast cancer patients. BMC Cancer. 2010; 10:247.
10. Jeschke J, O'Hagan HM, Zhang W, Vatapalli R, Calmon MF, Danilova L, et al. Frequent inactivation of cysteine dioxygenase type 1 contributes to survival of breast cancer cells and resistance to anthracyclines. Clin Cancer Res. 2013; 19:3201–3211.
11. Yang Y, Fan YC, Gao S, Dou CY, Zhang JJ, Sun FK, et al. Methylated cysteine dioxygenase-1 gene promoter in the serum is a potential biomarker for hepatitis B virus-related hepatocellular carcinoma. Tohoku J Exp Med. 2014; 232:187–194.
12. Kim BY, Suh KS, Lee JG, Woo SR, Park IC, Park SH, et al. Integrated analysis of prognostic gene expression profiles from hepatitis B virus-positive hepatocellular carcinoma and adjacent liver tissue. Ann Surg Oncol. 2012; 19:Suppl 3. S328–S338.
13. Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007; 8:286–298.
14. Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003; 349:2042–2054.
15. Tamura M, Gu J, Takino T, Yamada KM. Tumor suppressor PTEN inhibition of cell invasion, migration, and growth: differential involvement of focal adhesion kinase and p130Cas. Cancer Res. 1999; 59:442–449.
16. Lee S, Lee HJ, Kim JH, Lee HS, Jang JJ, Kang KH. Aberrant CpG island hypermethylation along multistep hepatocarcinogenesis. Am J Pathol. 2003; 163:1371–1378.
17. Nishida N, Nagasaka T, Nishimura T, Ikai I, Boland CR, Goel A. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology. 2008; 47:908–918.
18. Hu TH, Huang CC, Lin PR, Chang HW, Ger LP, Lin YW, et al. Expression and prognostic role of tumor suppressor gene PTEN/MMAC1/TEP1 in hepatocellular carcinoma. Cancer. 2003; 97:1929–1940.
19. Um TH, Kim H, Oh BK, Kim MS, Kim KS, Jung G, et al. Aberrant CpG island hypermethylation in dysplastic nodules and early HCC of hepatitis B virus-related human multistep hepatocarcinogenesis. J Hepatol. 2011; 54:939–947.
20. Tung-Ping Poon R, Fan ST, Wong J. Risk factors, prevention, and management of postoperative recurrence after resection of hepatocellular carcinoma. Ann Surg. 2000; 232:10–24.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download