Abstract
Objective
To evaluate the prognostic significance of p16, Ki-67, p63, and cytokeratin (CK) 17 expression determined by immunohistochemical staining in cervical intraepithelial neoplasia (CIN) 1.
Methods
Biopsy tissue samples from 33 patients diagnosed with CIN 1 were stained immunohistochemically for p16, Ki-67, p63, and CK17. The staining results were correlated with the clinical course of the disease.
Results
Seventeen of 18 (94.4%) p16-negative patients experienced regression, and only 1 patient (5.6%) developed persistent disease. Fifteen of the 16 (93.8%) Ki-67-negative patients experienced regression, and 1 patient (6.3%) developed persistent disease. Negative p16 and Ki-67 expression correlated significantly with disease regression (P=0.004 and P=0.017, respectively). Fourteen of 15 (93.3%) patients negative for both p16 and Ki-67 experienced regression, and 1 patient negative for both p16 and Ki-67 (6.7%) developed persistent disease. The expression levels of p63 and CK17 were not significantly associated with disease regression or persistence (P=0.149 and P=0.642, respectively). Ten of the 13 (76.9%) p16-positive patients had a high-risk HPV infection. High-risk HPV infection was significantly associated with p16 expression (P=0.049).
REFERENCES
1. Ostör AG. Natural history of cervical intraepithelial neoplasia: a critical review. Int J Gynecol Pathol. 1993; 12:186–92.
2. Schlecht NF, Platt RW, Duarte-Franco E, Costa MC, Sobrinho JP, Prado JC, et al. Human papillomavirus infection and time to progression and regression of cervical intraepithelial neoplasia. J Natl Cancer Inst. 2003; 95:1336–43.

3. Nobbenhuis MA, Helmerhorst TJ, van den Brule AJ, Rozendaal L, Voorhorst FJ, Bezemer PD, et al. Cytological regression and clearance of high-risk human papillomavirus in women with an abnormal cervical smear. Lancet. 2001; 358:1782–3.

4. Wright TC Jr, Massad LS, Dunton CJ, Spitzer M, Wilkinson EJ, Solomon D, et al. 2006 consensus guidelines for the management of women with cervical intraepithelial neoplasia or adenocarcinoma in situ. Am J Obstet Gynecol. 2007; 197:340–5.

5. Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim Biophys Acta. 1998; 1378:F115–77.

6. Klaes R, Friedrich T, Spitkovsky D, Ridder R, Rudy W, Petry U, et al. Overexpression of p16 (INK4A) as a specific marker for dysplastic and neoplastic epithelial cells of the cervix uteri. Int J Cancer. 2001; 92:276–84.
7. Keating JT, Cviko A, Riethdorf S, Riethdorf L, Quade BJ, Sun D, et al. Ki-67, cyclin E, and p16INK4 are complimentary surrogate biomarkers for human papilloma virus-related cervical neoplasia. Am J Surg Pathol. 2001; 25:884–91.
8. Negri G, Egarter-Vigl E, Kasal A, Romano F, Haitel A, Mian C. p16INK4a is a useful marker for the diagnosis of adenocarcinoma of the cervix uteri and its precursors: an immunohistochemical study with immunocytochemical correlations. Am J Surg Pathol. 2003; 27:187–93.
9. O'Neill CJ, McCluggage WG. p16 expression in the female genital tract and its value in diagnosis. Adv Anat Pathol. 2006; 13:8–15.
10. Negri G, Vittadello F, Romano F, Kasal A, Rivasi F, Girlando S, et al. p16INK4a expression and progression risk of lowgrade intraepithelial neoplasia of the cervix uteri. Virchows Arch. 2004; 445:616–20.

11. Negri G, Bellisano G, Zannoni GF, Rivasi F, Kasal A, Vittadello F, et al. p16 ink4a and HPV L1 immunohistochemistry is helpful for estimating the behavior of lowgrade dysplastic lesions of the cervix uteri. Am J Surg Pathol. 2008; 32:1715–20.
12. Murphy N, Ring M, Killalea AG, Uhlmann V, O'Donovan M, Mulcahy F, et al. p16INK4A as a marker for cervical dyskaryosis: CIN and cGIN in cervical biopsies and ThinPrep smears. J Clin Pathol. 2003; 56:56–63.
13. al-Saleh W, Delvenne P, Greimers R, Fridman V, Doyen J, Boniver J. Assessment of Ki-67 antigen immunostaining in squamous intraepithelial lesions of the uterine cervix. Correlation with the histologic grade and human papillomavirus type. Am J Clin Pathol. 1995; 104:154–60.
14. Isacson C, Kessis TD, Hedrick L, Cho KR. Both cell proliferation and apoptosis increase with lesion grade in cervical neoplasia but do not correlate with human papillomavirus type. Cancer Res. 1996; 56:669–74.
15. Kruse AJ, Baak JP, de Bruin PC, Jiwa M, Snijders WP, Boodt PJ, et al. Ki-67 immunoquantitation in cervical intraepithelial neoplasia (CIN): a sensitive marker for grading. J Pathol. 2001; 193:48–54.

16. Kruse AJ, Baak JP, Janssen EA, Kjellevold KH, Fiane B, Lovslett K, et al. Ki67 predicts progression in early CIN: validation of a multivariate progression-risk model. Cell Oncol. 2004; 26:13–20.

17. Quade BJ, Yang A, Wang Y, Sun D, Park J, Sheets EE, et al. Expression of the p53 homologue p63 in early cervical neoplasia. Gynecol Oncol. 2001; 80:24–9.

18. Martens JE, Arends J, Van der Linden PJ, De Boer BA, Helmerhorst TJ. Cytokeratin 17 and p63 are markers of the HPV target cell, the cervical stem cell. Anticancer Res. 2004; 24:771–5.
19. Regauer S, Reich O. CK17 and p16 expression patterns distinguish (atypical) immature squamous metaplasia from high-grade cervical intraepithelial neoplasia (CIN III). Histopathology. 2007; 50:629–35.

20. Smedts F, Ramaekers FC, Vooijs PG. The dynamics of keratin expression in malignant transformation of cervical epithelium: a review. Obstet Gynecol. 1993; 82:465.
21. Smedts F, Ramaekers F, Troyanovsky S, Pruszczynski M, Link M, Lane B, et al. Keratin expression in cervical cancer. Am J Pathol. 1992; 141:497–511.
Fig. 1.
(A) H&E, ×100, (B) H&E, ×200. The expression of p16 (C, D), Ki-67 (E, F), and p63 (G, H) in the lower one third of the cervical epithelium (×100, ×200). Note the loss of the CK17 expression in the lower one third of the cervical epithelium (I, J) ×100, ×200. (C-J) immunohistochemical stain.

Fig. 2.
The association of positive P16, Ki-67, p63, and CK17 immunohistochemical expressions with high risk HPV infection. aPearson's chi-square test, bFisher's exact test.
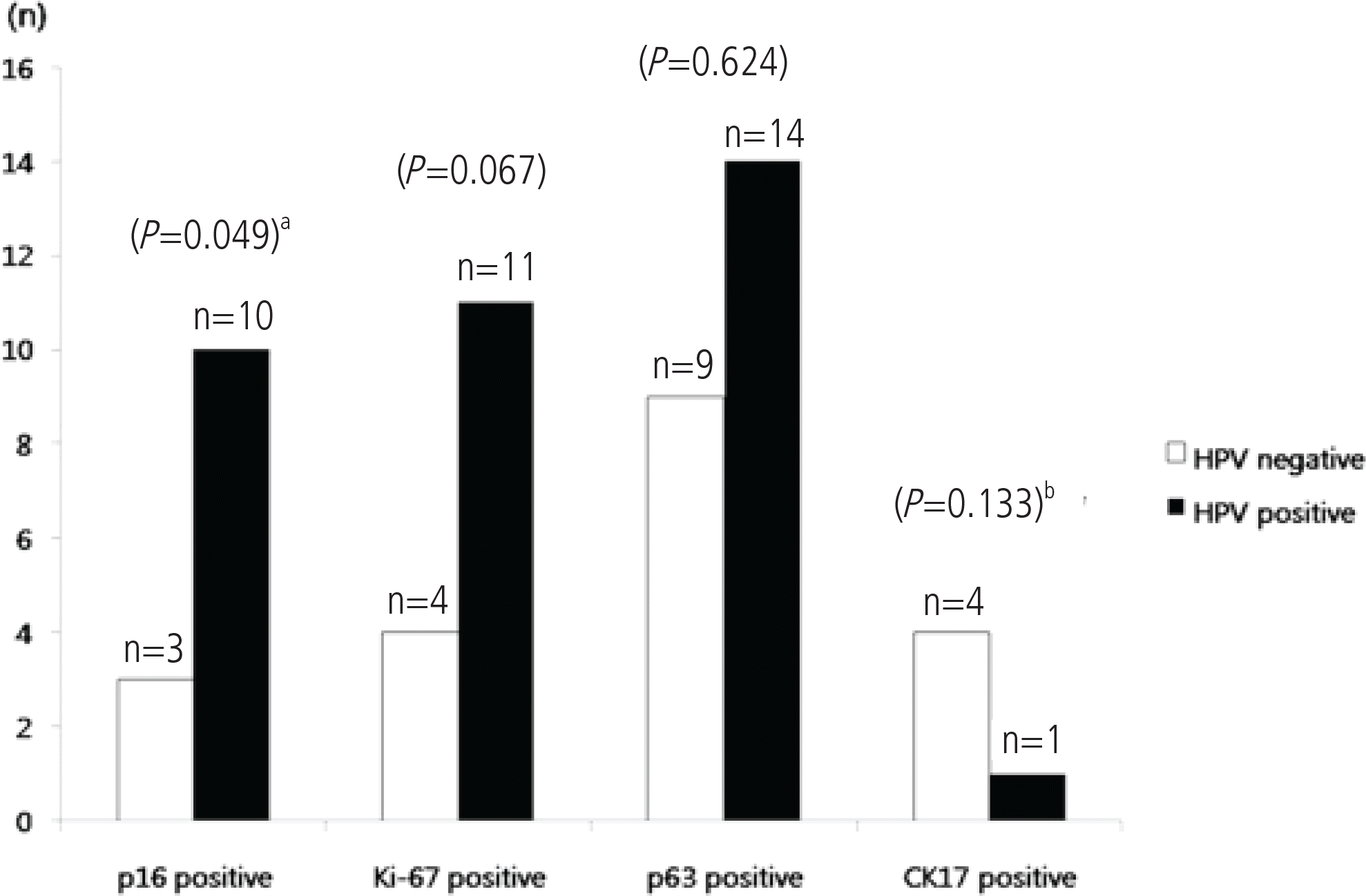
Table 1.
The behavior of CIN 1 and the expressions of p16, Ki-67, p63, and CK17
Table 2.
The behavior of CIN 1 as related to the expression statuses of p16, Ki-67, p63 and CK17
| Behavior | Total (n) | p16 | Ki‐67 | p63 | CK17 | p16/Ki‐67 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | + | - | + | - | + | - | + | -/- | -/+, +- | +/+ | ||
| Regression | 24 | 17 (94.4) | 7 (46.7) | 15 (93.8) | 9 (52.9) | 7 (100) | 17 (65.4) | 18 (69.2) | 6 (85.7) | 14 (93.3) | 4 (100) | 6 (42.9) |
| Persistence | 9 | 1 (5.6) | 8 (53.3) | 1 (6.3) | 8 (47.1) | 0 | 9 (34.6) | 8 (30.8) | 1 (14.3) | 1 (6.7) | 0 | 8 (57.1) |
| Total | 33 | 18 (54.6) | 15 (45.4) | 16 (48.5) | 17 (51.5) | 7 (21.2) | 26 (78.8) | 26 (78.8) | 7 (21.2) | 15 (45.4) | 4 (12.1) | 14 (42.4) |
| P-valuea | 0.004 | 0.017 | 0.149 | 0.642 | ||||||||




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download