Abstract
Bladder cancer (BC) is the fifth most common cancer worldwide and is associated with significant morbidity and mortality. The prognosis of muscle invasive BC is poor, and recurrence is common after radical surgery or chemotherapy. Therefore, new diagnostic methods and treatment modalities are critical. MicroRNAs (miRNAs), a class of small noncoding RNAs, regulate the expression of protein-coding genes by repressing translation or cleaving RNA transcripts in a sequence-specific manner. miRNAs have important roles in the regulation of genes involved in cancer development, progression, and metastasis. The availability of genomewide miRNA expression profiles by deep sequencing technology has facilitated rapid and precise identification of aberrant miRNA expression in BC. Indeed, several miRNAs that are either upregulated or downregulated have been shown to have associations with significant cancer pathways. Furthermore, many miRNAs, including those that can be detected in urine and blood, have been studied as potential noninvasive tumor markers for diagnostic and prognostic purposes. Here, we searched PubMed for publications describing the role of miRNAs in BC by using the keywords "bladder cancer" and "microRNA" on March 1, 2016. We found 374 papers and selected articles written in English in which the level of scientific detail and reporting were sufficient and in which novel findings were demonstrated. In this review, we summarize these studies from the point of view of miRNA-related molecular networks (specific miRNAs and their targets) and miRNAs as tumor markers in BC. We also discuss future directions of miRNA studies in the context of therapeutic modalities.
Bladder cancer (BC) is one of the leading causes of cancer-related death worldwide, with an estimated 429,800 new cases and 165,100 deaths globally in 2012 [1]. BCs can be categorized into 2 groups: nonmuscle invasive BC (NMIBC) and muscle invasive BC (MIBC). Approximately 70% to 80% of patients are diagnosed with NMIBC, and some of these patients have a high risk of recurrence and a variable risk of progression despite administration of local therapies [2]. The remaining 25% of patients with MIBC are managed by radical surgery or radiotherapy, but often still have poor outcomes, despite administration of these systemic therapies [2]. For patients with advanced BC or failure of first-line chemotherapy, no clearly defined second-line treatments have been shown to prolong overall survival [2]. Therefore, elucidation of novel molecular mechanisms of BC progression and identification of novel tumor markers that can predict recurrence or survival are urgently needed.
Despite its relatively low sensitivity (30%–40%), urinary cytology is a reliable marker for BC diagnosis because of its high specificity (90%–95%). However, because of the low sensitivity of urinary cytology, patients are still forced to undergo a painful cystoscopic procedure to confirm the initial diagnosis and to check for recurrence during follow-up examinations after undergoing transurethral resection of bladder tumors. Several new urine-based tests for BC, such as bladder tumor antigen, nuclear matrix protein 22 (NMP22, Sysmex Corp., Kobe, Japan), and FISH (UroVysion, Abbott Molecular Inc, Des Plaines, IL, USA), have been approved for clinical use. However, these new urinary markers have not been widely applied because they have relatively low specificities (60%–80%) compared with urinary cytology, despite their higher sensitivities (50%–70%) [3]. Thus, specificity may come at the cost of sensitivity, and conventional urinary cytology is a good example of this compromise. Accordingly, new effective diagnostic markers are urgently needed for patients with BC.
MicroRNAs (miRNAs) are endogenous small noncoding RNA molecules (19–22 bases in length) that negatively regulate the expression of protein-coding genes in a sequence-specific manner [4]. The nature of miRNAs is unique in that one miRNA can regulate multiple protein-coding RNAs. Bioinformatics predictions have indicated that miRNAs regulate 30% to 60% of the protein-coding genes in the human genome [56]. Growing evidence has demonstrated that aberrantly expressed miRNAs can act as oncogenes or tumor suppressors in various types of malignancies [7]. These miRNAs can disrupt tightly controlled RNA networks in cancer cells [8]. Since 2006, we have rigorously investigated the molecular networks of miRNAs and their target genes in clinical BC. Identification of aberrantly expressed miRNAs in BC could provide important clues for the investigation of novel molecular mechanisms of initiation, progression, and metastasis in BC. In this review, we discuss the role of miRNAs in BC on the basis of up-to-date results from our laboratory and other research groups.
After the completion of the Human Genome Project in 2003, about 20,000 to 25,000 genes were identified, and the sequences of 3 billion base pairs of human DNA were determined [9]. Surprisingly, only 2% of the human genome encodes functional proteins; 50% to 75% of the human genome is transcribed, and 98% of the transcripts are not translated into protein. Fig. 1 shows the current consensus of epigenetic regulation by noncoding RNAs transcribed from approximately 70% of the genomic region that used to be considered as "junk DNA" [10]. These noncoding RNAs can be roughly classified into two groups according to size. The first group includes short RNAs of less than 200 nucleotides (nt) in length; miRNAs, which are small RNA molecules of around 18 to 22 nt in length, fit into this group, as do other classes of small RNAs, such as piwi-interacting RNAs (around 23–30 nt) [11]. The other group includes long noncoding RNAs (lncRNAs) of around 200 nt or more. lncRNAs have been implicated in a range of developmental processes and diseases and can inhibit or activate gene expression through a range of diverse mechanisms. In contrast to miRNAs, which have been extensively studied and have well-understood functions in gene regulation, lncRNAs are not well studied or characterized.
miRNAs are evolutionarily conserved and are located either within the introns or exons of protein-coding genes (70%) or in intergenic regions (30%). Most intronic and exonic miRNAs are derived from their host genes, which suggests that they are transcribed concurrently with their host transcripts. Transcripts containing primary miRNAs (pri-miRNAs), which can vary from 200 nt to several kb in length, are capped with a specially modified nucleotide at the 5'-terminus and are polyadenylated with multiple adenosines at the 3'-end. Pri-miRNA is cleaved into precursor-miRNA (pre-miRNA: 60–70 nt in length) by the RNAse III enzyme (also known as Drosha). Pre-miRNA is exported from the nucleus into the cytoplasm by exportin-5. In the cytoplasm, pre-miRNA is cleaved by another RNase III enzyme, known as Dicer, into miRNA duplexes of approximately 19–22 nt in length. One miRNA duplex is then recruited into the RNA-induced silencing complex and functions to recognize complementary sites within the target messenger RNA (mRNA), thereby regulating translation through mRNA cleavage, degradation, or transcriptional repression [12].
miRNA expression profiling by array-based and polymerase chain reaction (PCR)-based methods is not commonly used because these methods only allow for identification of a limited number of known miRNAs. The rapid development of high-throughput, deep sequencing (DS) technology has provided novel information regarding the functions and features of miRNAs. Han et al. [13] first reported miRNA profiling in BC by using an Illumina Genome Analyzer IIX in 2011. Soon thereafter, five studies used DS technology to establish the miRNA profiles of clinical BC samples [1415161718]. To date, the largest DS study on BC is a comprehensive investigation by The Cancer Genome Atlas Research Network [18]. Tissues from 131 patients with high-grade MIBC were analyzed by RNA sequencing together with 118 blood samples from the same individuals and 23 adjacent histologically proven normal tissues. The aim of the project was to carry out genetic and epigenetic characterization of the molecular landscape of high-grade MIBC. In our studies, we conducted miRNA profiling by use of both PCRbased methods and DS technology [1719]. When comparing our 2 studies, the altered expression of miRNAs detected by DS technology was very similar to the results of PCRbased methods. In addition, a considerable number of novel miRNAs have been reported by some studies [131417]. Therefore, this analysis supports that DS technology is superior to previously used methods. Although PCR-based and DS profiles are not comparable from a quantitative point of view, sequencing approaches should provide substantial improvements to transcriptome analyses.
Growing evidence has demonstrated that aberrantly expressed miRNAs can act as oncogenic miRNAs (oncomiRNAs) or tumor-suppressive miRNAs (TS-miRNAs) in BC [1220]. Because miRNAs can construct miRNA-target gene networks that contribute to tumor initiation, survival, and invasion, many investigators have focused on the genes targeted by aberrantly expressed miRNAs in BC in comparison with normal bladder epithelium. In terms of miRNA analysis, luciferase-reporter assays are useful for confirming the direct binding of miRNAs to the specific 3'-untranslated region of the mRNA representing the target gene. Hence, we have summarized the miRNA studies in BC for which the target genes were validated by luciferase-reporter assays. To date, we have identified 78 studies that satisfied our criteria (Tables 1 and 2). As shown in Table 1, several TS-miRNAs, including miR-1, miR-16, miR-24, miR-101, miR-125b, miR-129, and miR-133a, have been shown to be located on different chromosomal regions but to have common sequences in their mature miRNAs. This suggests that the expression of these TS-miRNAs may be maintained by another genomic region, even though one region may be functionally disordered because of methylation, deletion, or mutation. This type of backup system is thought to have important roles in protecting normal cells from carcinogenesis.
The miRNAs in Table 1 are listed in order of miRNA number, and the validated target genes are indicated. Interestingly, among the 54 miRNAs examined, 45 were downregulated, whereas 9 were upregulated. In our experience, we often encountered difficulties with the use of anti-miRNAs in in vitro studies; however, miRNA restoration systems using miRNA transfection are well established and commercially available. Accordingly, the latter system may be easier to use to examine the potential tumor-suppressive roles of the downregulated miRNAs.
miRNAs have been shown to regulate target gene expression. Several miRNAs and their targets have been identified in BC (Table 2). Among the 61 target genes examined, the expression levels in clinical BC specimens compared with the levels in normal bladder epithelium were validated for 40 genes. The downregulation of several miRNAs, including miR-1, miR-133a, miR-145, miR-195, and the miR-200 family, has been reported, and these miRNAs have been shown to be involved in crucial cellular processes, such as the epithelial-mesenchymal transition (EMT), cell cycle, apoptosis, and cancer signaling pathways. TS-miRNAs and their target genes were classified into functional annotations by using the Kyoto Encyclopedia of Genes and Genomes pathways (Table 2).
Several miRNAs are located in close proximity (within 10 kbp) in the same genomic region; this is called a miRNA cluster. Because of their simultaneous expression, common target genes of miRNAs within a cluster may be important for tumor biology. For example, miR-145 forms a cluster with miR-143 on chromosome 5q32, and these clustered miRNAs are regulated by a common promoter [21]. Research indicates that both miR-145 and miR-143 are frequently downregulated in a broad range of cancers, including BC. Villadsen et al. [22] suggested that SERPINE1 is a direct target of both miR-145 and miR-143; this was the first report of an miRNA cluster targeting the same mRNA in BC. Importantly, miR-145 has been implicated in the p53 regulatory network in human malignancies [23]. p53 transcriptionally induces the expression of miR-145 by interaction with a potential p53 response element in the miR-145 promoter; moreover, MYC is directly repressed by miR-145. Additionally, Chiyomaru et al. [24] found direct binding of the miR-23b/27b cluster with EGFR/MET, and Itesako et al. [17] found direct binding of the miR-195/497 cluster with BIRC5, which encodes survivin.
Avoidance of apoptosis and cell cycle acceleration are required for cancer cells to escape cell death. BCL2L2, a member of the BCL2 family, can promote cell survival by inhibiting the intrinsic pathway of apoptosis. In contrast, BIRC5 (survivin) is a key member of the inhibitor of apoptosis protein family. Several TS-miRNAs induce apoptosis through targeting BCL2L2 (miR-133b, miR-203) and BIRC5 (miR-195, miR-203, miR-497). Upregulation of miR-129 may be involved in avoidance of apoptosis through direct targeting of SOX4 and GALNT1, which are involved in transcription and protein expression, respectively [25]. One important pathway is the sequential activation of caspases, which have a central role in the execution of cell apoptosis. miR-1 is downregulated in both clinical BC tissues and cultured BC cells and has been shown to induce apoptosis by increasing the activity of caspases 3 and 7 through direct targeting of the apoptosis inhibitor SRSF9 [26]. Additionally, CASP2 (caspase 2) is directly targeted by an onco-miRNA, miR-706 [27].
Several miRNAs have been shown to control the cell cycle through their target genes. Matsushita et al. [28] first reported that cell-cycle-related genes, such as CCNE1/2, CDC25A, and PKMYT1, are directly regulated by miR-144-5p. CDC25A is also regulated by miR-449a, and CDK4 and CDK6 are regulated by miR-124-3p, miR-195, miR-29c, miR-230c, and miR-449a, inducing G1-phase arrest. Interestingly, miR-96, which is upregulated in BC, directly binds to CDKN1A (p21), a well-known tumor-suppressive gene [29]. Moreover, miR-125b and miR-200c target the oncogene E2F3, which is critical for the G1/S transition and is overexpressed in most high-grade BCs [3031]. Inoguchi et al. [32] found that miR-24 regulates FOXM1, which is critical for mediating cell cycle progression proteins, such as Aurora B, cyclin B1, and CDC25B, and stimulates the expression of SKP2/CKS1, which is involved in the proteolysis of p27kip1 and in G1/S progression.
We also evaluated whether the expression of the different target genes could be validated in clinical BC samples (Table 2). The EMT is a key progression mechanism that is often activated during cancer invasion and metastasis and is characterized by loss of cell adhesion, repression of E cadherin expression, and increased cell mobility [33]. Members of the miR-200 family are well-known regulators of the EMT in human malignancies, including BC [34], and some investigators have reported the functions of these miRNAs in BC [3536].
FSCN1 and LASP1 are components of the filopodia and lamellipodia in various types of cells; both proteins are involved in mediating the dynamics of actin filaments and are activated during the EMT. miR-133a and miR-145 directly regulate FSCN1 [37], and the miR-1/133a cluster and miR-218 regulate LASP1 [38]. Restoration of the expression of these miRNAs accelerates cell migration and invasion activity in vitro [3738]. Matrix metalloproteinases (MMPs) are calcium-dependent zinc-containing endopeptidases that are capable of degrading various extracellular matrix proteins. MMPs are involved in the cleavage of cell surface receptors, the release of apoptotic ligands (such as the FAS ligand), and chemokine/cytokine inactivation. MMPs are also thought to facilitate tumor cell invasion and metastasis via the degradation of the extracellular matrix. Transforming growth factor beta1 reduces the expression level of miR-200b, which directly regulates MMP16. This then activates other MMPs (e.g., MMP-2 and MMP-9), growth factors, and receptors, thereby facilitating local cellular mechanisms that promote migration [39].
Loss of expression of miR-23b, miR-138, and miR-200c (an miR-200 family member) is associated with increased expression of ZEB1, ZEB2, and ERFFI1, which facilitate the progression of the EMT [364041]. ZEB1 and ZEB2 are known to negatively regulate E cadherin expression and are directly targeted by miR-23b and miR-138, whereas ERRFI-1, a regulator of EGFR, is directly targeted by members of the miR-200 family [36]. ZEB1 expression has been reported in 22% of clinical BC tissue specimens, but is absent from the bladder mucosa [42]. Expression of the miR-200 family has been shown to be repressed by hypermethylation of its promoter region, and loss of miR-200c expression is significantly associated with subsequent disease progression to MIBC and poor outcomes [35]. These studies implied that many EMT-related genes are activated through downregulation of several TS-miRNAs, and these phenomena may contribute to tumor progression of BC.
Downregulated miRNAs have also been reported to target some genes related to cytokine–cytokine receptor interactions, including EGFR (miR-23b, miR-27b, miR-133a, miR-133b) [2443], IGF1R (miR-145) [44], and MET (miR-23b, miR-27b, miR-101) [2443]. Activation of these genes may affect downstream cancer pathways, such as mammalian target of rapamycin signaling, phosphoinositol 3-kinase (PI3K)-Akt signaling, and mitogen-activated protein kinase (MAPK) signaling. FGFR3, which belongs to the MAPK signaling pathway, is crucial for BC oncogenesis, and mutations in FGFR3 have been shown to accelerate proliferation in BC [45]. Downregulation of miR-99a and miR-100 has been shown to cause upregulation of FGFR3 expression before its mutation, which suggests that the acquisition of mutations may result from increased cell turnover [46]. The expression of AKT1 and ILK, which are involved in the AKT-PI3K pathway, was shown to be repressed by the miR-143/145 cluster, and proliferation is inhibited in cells transfected with these miRNAs [4748]. The vascular endothelial growth factor (VEGF) and WNT signaling pathways are also important for BC development, and VEGFC, FZD4, PSEN1, and WNT1 have been reported to be under the control of several TS-miRNAs [1749505152].
Many studies have investigated miRNA expression by quantitative real-time PCR of clinical BC specimens [20]. In this review, we focus on studies in which the investigators evaluated diagnostic accuracy by use of receiver-operator curve analyses of the sensitivity and specificity of specific miRNAs in comparison with normal bladder epithelium. We found 18 studies that satisfied our criteria (Table 3). In 2009, Ichimi et al. [19] first evaluated the expression of six miRNAs in tissues as diagnostic markers to distinguish BC (n=104) from non-BC (n=31).
There is a pressing need for highly accurate, noninvasive tests for BC diagnosis. The standard procedures for BC diagnosis and the follow-up for patients depend on urinary cytology and cystoscopy. Urinary cytology is a reliable urine marker for BC diagnosis because of its high specificity (90%–95%). However, this method has low sensitivity (30%–40%), and patients are forced to undergo a painful cystoscopic procedure to confirm the diagnosis. A number of noninvasive urine tests, such as bladder tumor antigen and NMP22, have been developed; however, their diagnostic specificities are inferior to that of urinary cytology tests, and these methods have failed to meet clinical expectations. Therefore, quantitative changes in miRNAs in urine, blood, and tissues are the primary focus in the search for new markers. miRNAs are detectable in urine with pelleted or cell-free urine samples. Hanke et al. [53] evaluated miR-126 expression in whole urine samples and showed that this miRNA had a sensitivity of 72% and specificity of 82% when used as a diagnostic marker, even though the sample size in their study was small. Yamada et al. [54] also found that miR-96 detection in the urine including the pellet was a useful diagnostic marker, with a sensitivity of 71.8% and a specificity of 89.2%. Shimizu et al. [55] uniquely reported good sensitivity of 81% and good specificity of 89% by testing a methylation panel of miR-9-3/miR-124-2/miR-124-3/miR-137. In terms of miRNA detection in cell-free urine [5657585960], several studies have shown that this method provides moderate sensitivity and specificity for distinguishing BC from non-BC but that the results are inferior to those from studies using whole urine samples [5354556162]. In blood tests, Jiang et al. [63] showed dramatic results (sensitivity of 90% and specificity of 90%) using a panel study of miR-15b-5p/miR-27a-3p/miR-30a-5p/miR-148b-3p/miR-152/miR-3187-3p.
Many retrospective studies have suggested that the expression of specific miRNAs in BC specimens may be a good prognostic marker predicting overall survival, disease-free survival, recurrence-free survival, or progression-free survival, as outlined in Table 4. Moreover, several reports analyzing urine and blood samples have shown that miR-200, miR-214, and miR-3187-3p levels are good prognostic markers in the blood [57606364]. Surprisingly, miR-203, miR-214, miR-152, and miR-3187-3p were shown to be independent markers predicting overall survival, progression-free survival, or recurrence-free survival in multivariate studies [636465]. Unfortunately, no prospective studies have been performed to date. Thus, additional large, prospective studies are needed for the clinical application of miRNA assays for diagnostic and prognostic purposes in BC.
Some TS-miRNAs have been transfected into BC cell lines by using plasmid/virus vectors, followed by subcutaneous injection of the transfected cells into nude mice [276667]. Significant inhibition of tumor growth [2767] and metastasis [66] was observed. Yu et al. [66] found that the CD44 gene was a direct target of miR-34a and that the pro-apoptotic role of miR-34a may be mediated primarily through regulation of CD44, which functions to promote metastasis and angiogenesis. Xu et al. [68] demonstrated that miR-100-transfected BC cells exhibited significant growth inhibition in an intravesical orthotopic BC model. In terms of onco-miRNAs, miR-10b- and miR-155-transfected BC xenografts exhibited significantly increased growth in comparison with controls [6970]. Inamoto et al. [71] tested the anticancer effects of intravesical injection of miR-145 by using cationic liposomes (Lipofectamine RNAiMAX, Thermo Fisher Scientific, Waltham, MA, USA) in an intravesical orthotopic BC model. They achieved adequate anticancer effects after intravesical administration of miR-145. These miRNAs are promising candidate miRNAs that may be useful for clinical applications in the future.
Many studies have examined the roles of miRNAs in human malignancies, including BC. However, the majority of miRNA studies have focused on candidate gene approaches with limited whole-genome sequencing, and a detailed characterization of miRNA profiles based on the genomic features of BC has not been reported. Recently, DS technology has been applied for profiling of mRNA and miRNA in human cancers, including BC. However, DS technology has mainly focused on BC tissues without any particular selection according to clinical treatment. Additional DS analyses should be conducted using samples from patients who have undergone different treatments, e.g., chemotherapy regimens, radiation therapies, or immune-checkpoint drugs.
A growing body of evidence has shown that miRNAs have great potential for clinical use as new diagnostic biomarkers. However, detection of single miRNAs in urine samples will not be superior to current urinary cytology tests because of the relatively low diagnostic specificity of such analyses. Some recent studies have attempted to improve the diagnostic specificity of miRNA detection by using a panel of multiple miRNAs [55566163]. It will be important to select optimal combinations of miRNAs and to perform prospective translational studies in outpatients to validate such miRNA panels.
Moreover, recent studies have shown that some miRNAs control the activity of major cancer-related signaling molecules. Thus, identification of aberrant miRNA expression and oncogenic or tumor-suppressive molecular targets of miRNAs is necessary for the clinical development of novel cancer therapeutics. Because many TS-miRNAs and their target oncogenes are components of complex molecular networks, treatment with multiple miRNAs may provide stronger anticancer effects than treatment with a single miRNA. In an interesting study by Liu et al. [72], new vectors, termed "miRNA-mowers," that contained the entire sequence of the onco-miR-183-96-182 cluster were constructed. Transfection of the miRNA-mowers strongly inhibited cell growth and migration and induced apoptosis in vitro, suggesting the usefulness of targeting multiple onco-miRNAs. However, no studies have yet reported successful results for intravenous administration of TS-miRNAs in the induction of anticancer effects in vivo. The development of an adequate drug delivery system for TS-miRNAs is needed; otherwise, it will be difficult to determine the value of miRNAs as a therapeutic modality.
Figures and Tables
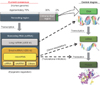 | Fig. 1Current consensus of epigenetic gene regulation by noncoding RNAs. microRNAs is classified as one of small noncoding RNAs, and epigenetically function as negative regulator of protein coding genes by complementary binding to the target messengerRNAs (mRNAs). ncRNA, noncoding RNA. |
Table 1
miRNAs and their target genes validated by lusiferase reporter assay

| miRNAs | Stem-loop sequence | Locus | Clustered miRNAs | miRNA type | Target | Reference |
|---|---|---|---|---|---|---|
| miR-1 | miR-1-1 | 20q13 | ND | TS | LASP1 | 38 |
| miR-1-2 | 18q11 | miR-133a-1 | PNP | 73 | ||
| PTMA | 73 | |||||
| SRSF9 | 26 | |||||
| TAGLN2 | 74 | |||||
| miR-16 | miR-16-1 | 13q14 | miR-15a | TS | CCND1 | 75 |
| miR-16-2 | 3q25 | miR-15b | ||||
| miR-23b | miR-23b | 9q22 | miR-27b | TS | EGFR | 24 |
| miR-3074 | MET | 24 | ||||
| miR-24-1 | ZEB1 | 40 | ||||
| miR-24 | miR-24-1 | 9q22 | miR-23b, miR-27b, miR-3074 | TS | CARD10 | 76 |
| miR-24-2 | 19p13 | miR-23a, miR-27a | FOXM1 | 32 | ||
| miR-27a | miR-27a | 19p13 | miR-23a, miR-24-2 | TS | RUNX1 | 77 |
| SLC7A11 | 78 | |||||
| miR-27b | miR-27b | 9q22 | miR-23b, miR-24-1, miR-3074 | TS | DROSHA | 79 |
| EGFR | 24 | |||||
| MET | 24 | |||||
| miR-29c | miR-29c | 1q32 | miR-29b-2 | TS | CDK6 | 80 |
| miR-30a | miR-30a | 6q13 | ND | TS | NOTCH1 | 81 |
| miR-34a | miR-34a | 1p36 | ND | TS | CD44 | 66 |
| HNF4G | 82 | |||||
| NOTCH1 | 83 | |||||
| miR-99a | miR-99a | 21q21 | let-7c | TS | FGFR3 | 46 |
| miR-100 | miR-100 | 11q24 | ND | TS | MTOR | 68 |
| miR-101 | miR-101-1 | 1p31 | miR-3671 | TS | COX2 | 84 |
| miR-101-2 | 9p24 | ND | MET | 85 | ||
| VEGFC | 52 | |||||
| miR-124-3p | miR-124 | 20q13 | ND | TS | ROCK1 | 86 |
| CDK4 | 87 | |||||
| miR-125b | miR-125b-1 | 11q24 | ND | TS | E2F3 | 31 |
| miR-125b-2 | 21q21 | ND | MMP13 | 88 | ||
| SPHK1 | 89 | |||||
| miR-128 | miR-128-1 | 2q21 | ND | TS | VEGFC | 49 |
| miR-128-2 | 3p22 | ND | ||||
| miR-129 | miR-129-1 | 7q32 | ND | TS | GALNT1 | 25 |
| miR-129-2 | 11p11 | ND | SOX4 | |||
| miR-133a | miR-133a-1 | 18q11 | miR-1-2 | TS | EGFR | 43 |
| miR-133a-2 | 20q13 | ND | FSCN1 | 37 | ||
| GSTP1 | 90 | |||||
| LASP1 | 38 | |||||
| PNP | 73 | |||||
| PTMA | 73 | |||||
| TAGLN2 | 74 | |||||
| miR-133b | miR-133b | 6p12 | miR-206 | TS | AKT1 | 47 |
| BCL2L2 | 47 | |||||
| EGFR | 43 | |||||
| miR-135a | miR-135a-1 | 3p21 | ND | TS | FOXO1 | 91 |
| miR-135a-2 | 12q23 | ND | ||||
| miR-138 | miR-138-1 | 3p21 | ND | TS | ZEB2 | 41 |
| miR-138-2 | 16q13 | ND | ||||
| miR-143 | miR-143 | 5q32 | miR-145 | TS | SERPIN1 | 22 |
| AKT | 48 | |||||
| miR-144-5p/3p | miR-144 | 17q11 | miR-451a, miR-451b, miR-4732 | TS | CCNE1 | 28 |
| CCNE2 | ||||||
| CDC25A | ||||||
| PKMYT1 | ||||||
| miR-145 | miR-145 | 5q32 | miR-143 | TS | CBFB | 92 |
| CLINT1 | 92 | |||||
| FSCN1 | 37 | |||||
| ILK | 48 | |||||
| PAK1 | 93 | |||||
| PPP3CA | 92 | |||||
| SERPIN1 | 22 | |||||
| SOCS7 | 94 | |||||
| IGF1R | 44 | |||||
| miR-186 | miR-186 | 1p31 | ND | TS | HMGN5 | 95 |
| miR-193a-3p | miR-193a | 17q11 | ND | TS | LOXL4 | 96 |
| PSEN1 | 52 | |||||
| HOXC9 | 97 | |||||
| miR-195 | miR-195 | 17p13 | miR-497 | TS | BIRC5 | 17 |
| CDC42 | 98 | |||||
| CDK4 | 99 | |||||
| GLUT3 | 100 | |||||
| WNT7A | 17 | |||||
| miR-200b | miR-200b | 1p36 | miR-200a, miR-429 | TS | MMP16 | 39 |
| miR-200c | miR-200c | 12p13 | miR-141 | TS | BMI1 | 30 |
| E2F3 | ||||||
| miR-203 | miR-203 | 14q32 | miR-203b | TS | BCL2L2 | 65 |
| BIRC5 | ||||||
| miR-214 | miR-214 | 1q24 | miR-199a-2, miR-3120 | TS | PDRG1 | 101 |
| miR-218 | miR-218-1 | 4p15 | ND | TS | BMI1 | 67 |
| miR-218-2 | 5q34 | ND | LASP1 | 38 | ||
| miR-221 | miR-221 | Xp11 | miR-222 | TS | STMN1 | 102 |
| miR-320a | miR-320a | 8p21 | ND | TS | ITGB3 | 103 |
| miR-320c | miR-320c | 18q11 | ND | TS | CDK6 | 104 |
| miR-449a | miR-449a | 5q11.2 | miR-449b, miR-449c | TS | CDC25A | 105 |
| miR-485-5p | miR-485 | 14q32 | ND | TS | HMGA2 | 106 |
| miR-490-5p | miR-490 | 7q33 | ND | TS | FOS | 107 |
| miR-493 | miR-493 | 14q32 | miR-337, miR-665 | TS | FZD4 | 51 |
| RHOC | ||||||
| miR-497 | miR-497 | 17p13 | miR-195 | TS | BIRC5 | 17 |
| WNT7A | ||||||
| miR-574-3p | miR-574 | 4p14 | ND | TS | MESDC1 | 108 |
| miR-576-3p | miR-576 | 4q25 | ND | TS | CCND1 | 109 |
| miR-590-3p | miR-590 | 7q11 | ND | TS | TFAM | 110 |
| miR-1182 | miR-1182 | 1q42 | ND | TS | TERT | 111 |
| miR-9 | miR-9-1 | 1q22 | ND | Onco | CBX7 | 112 |
| miR-9-2 | 5q14 | ND | ||||
| miR-9-3 | 15q26 | ND | CERS2 | 113 | ||
| miR-10b | miR-10b | 2q31 | ND | Onco | HOXD10 | 70 |
| KLF4 | ||||||
| miR-19a | miR-19a | 13q31 | miR-17, miR-18a, miR-20a, miR-19b-1, miR-92a-1 | Onco | PTEN | 114 |
| miR-96 | miR-96 | 7q32 | miR-18, miR-183 | Onco | CDKN1A | 29 |
| miR-150 | miR-150 | 19q13 | ND | Onco | PDCD4 | 115 |
| miR-155 | miR-155 | 21q21 | ND | Onco | DMTF1 | 69 |
| miR-182-5p | miR-182 | 7q32 | ND | Onco | RECK | 116 |
| SMAD4 | ||||||
| miR-708 | miR-708 | 11q14 | ND | Onco | CASP2 | 27 |
Table 2
Functional annotation of the target genes and the relative miRNAs

| Annotation | Target | miRNA | miRNA type | Target validation in clinical BCs | Reference |
|---|---|---|---|---|---|
| Apoptosis | BCL2L2 | miR-133b, miR-203 | TS | yes | 47,65 |
| BIRC5 | miR-195, miR-203, miR-497 | TS | yes | 17,65 | |
| BMI1 | miR-200c, miR-218 | TS | no | 30,67 | |
| CASP2 | miR-708 | Onco | yes | 27 | |
| SOCS7 | miR-145 | TS | no | 94 | |
| SOX4 | miR-129 | TS | yes | 25 | |
| SRSF9 | miR-1 | TS | yes | 26 | |
| Cell cycle | CCND1 | miR-16, miR-576-3p | TS | yes | 75,109 |
| CCNE1 | miR-144-5p/3p | TS | yes | 28 | |
| CCNE2 | miR-144-5p/3p | TS | yes | 28 | |
| CDC25A | miR-144-5p/3p | TS | yes | 28 | |
| CDC25A | miR-449a | TS | yes | 105 | |
| CDK4 | miR-124-3p | TS | no | 87 | |
| CDK4 | miR-195 | TS | yes | 99 | |
| CDK6 | miR-29c, miR-320c | TS | yes | 80,104 | |
| CDK6 | miR-449a | TS | yes | 105 | |
| CDKN1A | miR-96 | Onco | yes | 29 | |
| E2F3 | MiR-125b, miR-200c | TS | yes | 30,31 | |
| FOXM1 | miR-24 | TS | yes | 32 | |
| PKMYT1 | miR-144-5p/3p | TS | yes | 28 | |
| STMN1 | miR-221 | TS | no | 102 | |
| EMT | CARD10 | miR-24 | TS | no | 76 |
| CBX7 | miR-9 | Onco | yes | 112 | |
| FSCN1 | miR-133a | TS | yes | 37 | |
| FSCN1 | miR-145 | TS | yes | 37 | |
| HMGA2 | miR-485-5p | TS | no | 106 | |
| HMGN5 | miR-186 | TS | yes | 95 | |
| LASP1 | miR-1, miR-133a, miR-218 | TS | yes | 38 | |
| LOXL4 | miR-193a-3p | TS | no | 96 | |
| MESDC1 | miR-574-3p | TS | no | 108 | |
| MMP13 | MiR-125b | TS | no | 88 | |
| MMP16 | miR-200b | TS | no | 39 | |
| ZEB1 | miR-23b | TS | yes | 40 | |
| ZEB2 | miR-138 | TS | yes | 41 | |
| Cytokaine-cytokaine receptor intaraction | EGFR | miR-23b, miR-27b, miR-133a, miR-133b | TS | yes | 24,85 |
| IGF1R | miR-145 | TS | no | 44 | |
| MET | miR-23b, miR-27b, miR-101 | TS | yes | 24,85 | |
| MAPK signaling | FGFR3 | miR-99a, miR-100 | TS | yes | 46 |
| FOS | miR-490-5p | TS | yes | 108 | |
| RECK | miR-182-5p | Onco | yes | 116 | |
| Notch signaling | NOTCH1 | miR-30a, miR-34a | TS | yes | 81,83 |
| PI3K-Akt signaling | AKT1 | miR-133b | TS | yes | 47 |
| AKT1 | miR-143 | TS | no | 48 | |
| ILK | miR-145 | TS | no | 48 | |
| MTOR | miR-100 | TS | yes | 68 | |
| PTEN | miR-19a | Onco | no | 114 | |
| Ras signaling | CDC42 | miR-195 | TS | yes | 98 |
| PAK1 | miR-145 | TS | no | 93 | |
| RHOC | miR-493 | TS | no | 51 | |
| ROCK1 | miR-124-3p | TS | yes | 86 | |
| VEGF signaling | VEGFC | miR-101 | TS | yes | 49,52 |
| WNT signaling | FZD4 | miR-493 | TS | no | 51 |
| PSEN1 | miR-193a-3p | TS | no | 52 | |
| WNT7A | miR-195, miR-497 | TS | yes | 17 | |
| Metabolic | CERS2 | miR-9 | Onco | yes | 113 |
| COX2 | miR-101 | TS | no | 84 | |
| GALNT1 | miR-129 | TS | yes | 25 | |
| ECM-receptor | CD44 | miR-34a | TS | yes | 66 |
| ITGB3 | miR-320a | TS | yes | 103 |
Table 3
miRNAs as diagnostic markers

| Sample | Study | Year | Calculator | BC/Cont. (n) | Sensitivity | Specificity | Reference |
|---|---|---|---|---|---|---|---|
| Tissue | Liu | 2015 | miR-141 | 30/30 | 78.2 | 51.6 | 139 |
| miR-200b | 81.3 | 47 | |||||
| Ichimi | 2009 | miR-30a-3p | 104/31 | 94.3 | 76.7 | 19 | |
| miR-125 | 89.4 | 77.4 | |||||
| miR-133a | 93.3 | 77.4 | |||||
| miR-145 | 90.5 | 77.4 | |||||
| miR-195 | 91.4 | 80 | |||||
| miR-199a* | 72.1 | 90.3 | |||||
| Urine | Long | 2015 | miR-26a/miR-93/miR-191/miR-940 | 85/45 | 70 | 84 | 56 |
| Wang | 2015 | miR-214 | 192/169 | 90.5 | 65.6 | 57 | |
| Zhang | 2014 | miR-99a, miR-125b | 50 | 86.7 | 81.1 | 58 | |
| Zhou | 2014 | miR-106b | 112/78 | 76.8 | 72.4 | 59 | |
| Tolle | 2013 | miR-520e | 36/19 | 70 | 63.2 | 117 | |
| miR-618 | 70 | 68.4 | |||||
| miR-1255b-5p | 85 | 68.4 | |||||
| Mengual | 2013 | miR-18a*/miR-25/miR-140-5p/miR-142-3p/miR-187/miR-204 | 151/126 | 84.8 | 86.5 | 61 | |
| Shimizu | 2013 | miR-9-3/miR-124-2/miR-124-3/miR-137 | 86/20 | 81 | 89 | 55 | |
| Yun | 2012 | miR-145 | 207/144 | 77.8 | 61.1 | 60 | |
| Miah | 2012 | miR-15b/miR-135b/miR-1224-3p | 121/53 | 94.1 | 51 | 118 | |
| Dudziec | 2011 | miR-152/miR-328/miR-122-3p | 68/53 | 81 | 75 | 62 | |
| miR-212 | 40 | 66 | |||||
| miR-328 | 59 | 91 | |||||
| miR-1224-3p | 84 | 58 | |||||
| Yamada | 2011 | miR-96 | 100/74 | 71.8 | 89.2 | 54 | |
| miR-183 | 74 | 77.3 | |||||
| Hanke | 2010 | miR-126: miR-152 ratio | 29/11 | 72 | 82 | 53 | |
| miR-182: miR-152 ratio | 55 | 82 | |||||
| Blood | Motawi | 2016 | miR-92a | 70/62 | 97.1 | 76.7 | 119 |
| miR-100 | 90 | 66.7 | |||||
| miR-143 | 78.6 | 93.3 | |||||
| Fang | 2015 | miR-205 | 89/56 | 76.4 | 96.4 | 120 | |
| Du | 2015 | miR-497/miR-663b | 109/115 | 69.7 | 69.6 | 121 | |
| Jiang | 2015 | miR-15b-5p/miR-27a-3p/miR-30a-5p/miR-148b-3p/miR-152/miR-3187-3p | 110/110 | 90 | 90 | 63 | |
| Tolle | 2013 | miR-26b-5p | 38/20 | 65 | 94.1 | 117 | |
| miR-144-5p | 70 | 82.4 | |||||
| miR-374-5p | 60 | 94.1 |
Table 4
miRNAs as prognostic markers
| Sample | Study | Year | Calculator | BC (n) | Valuables | Multivariate | Reference |
|---|---|---|---|---|---|---|---|
| Tissue | Zhang | 2016 | miR-30a | 50 | OS, RFS | N/A | 81 |
| Xu | 2015 | let-7c, miR-99a, miR-125b-1, miR-193a | 202 | OS | N/A | 124 | |
| Zhang | 2015 | miR-21 | 53 | OS | N/A | 122 | |
| Wang | 2015 | miR-141 | 114 | DSS | N/A | 127 | |
| Avgeris | 2015 | miR-143, miR-145 | 133 | OS, PFS | N/A | 125 | |
| Zhang | 2015 | miR-203 | 108 | OS, PFS | Independent | 65 | |
| Wu | 2015 | miR-424 | 124 | OS, RFS | N/A | 123 | |
| Segersten | 2014 | miR-10a-5p | 86 | PFS | N/A | 129 | |
| Lin | 2014 | miR-26a | 126 | OS, DFS | N/A | 126 | |
| Zhang | 2014 | miR-222 | 97 | OS | N/A | 128 | |
| Pignot | 2013 | miR-9, miR-182, miR-200b | 166 | OS, RFS | N/A | 134 | |
| Majid | 2013 | miR-23b | 20 | OS | N/A | 40 | |
| Dyrskjot | 2009 | miR-29c, miR-129, miR-133b | 106 | PFS | N/A | 25 | |
| Rosenberg | 2013 | miR-29c* | 108 | PFS | N/A | 132 | |
| Wang | 2013 | miR-31 | 126 | OS, PFS | N/A | 131 | |
| Ratert | 2013 | miR-141, miR-205 | 40 | OS | N/A | 130 | |
| Pignot | 2012 | miR-19a, miR-20a, miR-92a | 71 | OS | N/A | 138 | |
| Zaravinos | 2012 | miR-21 | 77 | OS | N/A | 136 | |
| Wang | 2012 | miR-100 | 126 | OS, PFS | N/A | 133 | |
| Hirata | 2012 | miR-182-5p | 18 | OS | N/A | 116 | |
| Puerta-Gil | 2012 | miR-222, miR-143 | 37 | OS, DSS, RFS, PFS | N/A | 137 | |
| Majid | 2012 | miR-1280 | 21 | OS | N/A | 135 | |
| Urine | Dudziec | 2011 | miR-152 CpG, miR-1224 CpG | 68 | PFS | N/A | 62 |
| Yun | 2012 | miR-200 | 138 | RFS | N/A | 60 | |
| Wang | 2015 | miR-214 | 192 | OS, RFS | N/A | 57 | |
| Kim | 2013 | miR-214 | 138 | RFS | Independent | 64 | |
| Blood | Jiang | 2015 | miR-152, miR-3187-3p | 110 | RFS | Independent | 63 |
| Fang | 2015 | miR-205 | 89 | DSS | N/A | 120 |
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015; 65:87–108.
2. Kaufman DS, Shipley WU, Feldman AS. Bladder cancer. Lancet. 2009; 374:239–249.
3. Schmitz-Drager BJ, Todenhofer T, van Rhijn B, Pesch B, Hudson MA, Chandra A, et al. Considerations on the use of urine markers in the management of patients with low-/intermediate-risk non-muscle invasive bladder cancer. Urol Oncol. 2014; 32:1061–1068.
4. Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009; 136:642–655.
5. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008; 9:102–114.
6. Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008; 319:1785–1786.
7. Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013; 774:1–20.
8. Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. Lancet Oncol. 2012; 13:e249–e258.
9. International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004; 431:931–945.
10. Huttenhofer A, Schattner P, Polacek N. Non-coding RNAs: hope or hype? Trends Genet. 2005; 21:289–297.
11. Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007; 23:175–205.
12. Yoshino H, Seki N, Itesako T, Chiyomaru T, Nakagawa M, Enokida H. Aberrant expression of microRNAs in bladder cancer. Nat Rev Urol. 2013; 10:396–404.
13. Han Y, Chen J, Zhao X, Liang C, Wang Y, Sun L, et al. MicroRNA expression signatures of bladder cancer revealed by deep sequencing. PLoS One. 2011; 6:e18286.
14. Chen YH, Wang SQ, Wu XL, Shen M, Chen ZG, Chen XG, et al. Characterization of microRNAs expression profiling in one group of Chinese urothelial cell carcinoma identified by Solexa sequencing. Urol Oncol. 2013; 31:219–227.
15. Zhu J, Jiang Z, Gao F, Hu X, Zhou L, Chen J, et al. A systematic analysis on DNA methylation and the expression of both mRNA and microRNA in bladder cancer. PLoS One. 2011; 6:e28223.
16. Li X, Chen J, Hu X, Huang Y, Li Z, Zhou L, et al. Comparative mRNA and microRNA expression profiling of three genitourinary cancers reveals common hallmarks and cancer-specific molecular events. PLoS One. 2011; 6:e22570.
17. Itesako T, Seki N, Yoshino H, Chiyomaru T, Yamasaki T, Hidaka H, et al. The microRNA expression signature of bladder cancer by deep sequencing: the functional significance of the miR-195/497 cluster. PLoS One. 2014; 9:e84311.
18. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014; 507:315–322.
19. Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009; 125:345–352.
20. Matullo G, Naccarati A, Pardini B. MicroRNA expression profiling in bladder cancer: the challenge of next-generation sequencing in tissues and biofluids. Int J Cancer. 2016; 138:2334–2345.
21. Iio A, Nakagawa Y, Hirata I, Naoe T, Akao Y. Identification of non-coding RNAs embracing microRNA-143/145 cluster. Mol Cancer. 2010; 9:136.
22. Villadsen SB, Bramsen JB, Ostenfeld MS, Wiklund ED, Fristrup N, Gao S, et al. The miR-143/-145 cluster regulates plasminogen activator inhibitor-1 in bladder cancer. Br J Cancer. 2012; 106:366–374.
23. Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009; 460:529–533.
24. Chiyomaru T, Seki N, Inoguchi S, Ishihara T, Mataki H, Matsushita R, et al. Dual regulation of receptor tyrosine kinase genes EGFR and c-Met by the tumor-suppressive microRNA-23b/27b cluster in bladder cancer. Int J Oncol. 2015; 46:487–496.
25. Dyrskjot L, Ostenfeld MS, Bramsen JB, Silahtaroglu AN, Lamy P, Ramanathan R, et al. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009; 69:4851–4860.
26. Yoshino H, Enokida H, Chiyomaru T, Tatarano S, Hidaka H, Yamasaki T, et al. Tumor suppressive microRNA-1 mediated novel apoptosis pathways through direct inhibition of splicing factor serine/arginine-rich 9 (SRSF9/SRp30c) in bladder cancer. Biochem Biophys Res Commun. 2012; 417:588–593.
27. Song T, Zhang X, Zhang L, Dong J, Cai W, Gao J, et al. miR-708 promotes the development of bladder carcinoma via direct repression of Caspase-2. J Cancer Res Clin Oncol. 2013; 139:1189–1198.
28. Matsushita R, Seki N, Chiyomaru T, Inoguchi S, Ishihara T, Goto Y, et al. Tumour-suppressive microRNA-144-5p directly targets CCNE1/2 as potential prognostic markers in bladder cancer. Br J Cancer. 2015; 113:282–289.
29. Wu Z, Liu K, Wang Y, Xu Z, Meng J, Gu S. Upregulation of microRNA-96 and its oncogenic functions by targeting CDKN1A in bladder cancer. Cancer Cell Int. 2015; 15:107.
30. Liu L, Qiu M, Tan G, Liang Z, Qin Y, Chen L, et al. miR-200c inhibits invasion, migration and proliferation of bladder cancer cells through down-regulation of BMI-1 and E2F3. J Transl Med. 2014; 12:305.
31. Huang L, Luo J, Cai Q, Pan Q, Zeng H, Guo Z, et al. MicroRNA-125b suppresses the development of bladder cancer by targeting E2F3. Int J Cancer. 2011; 128:1758–1769.
32. Inoguchi S, Seki N, Chiyomaru T, Ishihara T, Matsushita R, Mataki H, et al. Tumour-suppressive microRNA-24-1 inhibits cancer cell proliferation through targeting FOXM1 in bladder cancer. FEBS Lett. 2014; 588:3170–3179.
33. Yun SJ, Kim WJ. Role of the epithelial-mesenchymal transition in bladder cancer: from prognosis to therapeutic target. Korean J Urol. 2013; 54:645–650.
34. Abba ML, Patil N, Leupold JH, Allgayer H. MicroRNA regulation of epithelial to mesenchymal transition. J Clin Med. 2016; 5:8.
35. Wiklund ED, Bramsen JB, Hulf T, Dyrskjot L, Ramanathan R, Hansen TB, et al. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int J Cancer. 2011; 128:1327–1334.
36. Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A, et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res. 2009; 15:5060–5072.
37. Chiyomaru T, Enokida H, Tatarano S, Kawahara K, Uchida Y, Nishiyama K, et al. miR-145 and miR-133a function as tumour suppressors and directly regulate FSCN1 expression in bladder cancer. Br J Cancer. 2010; 102:883–891.
38. Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Uchida Y, Kawahara K, et al. Functional role of LASP1 in cell viability and its regulation by microRNAs in bladder cancer. Urol Oncol. 2012; 30:434–443.
39. Chen MF, Zeng F, Qi L, Zu XB, Wang J, Liu LF, et al. Transforming growth factor β1 induces epithelial mesenchymal transition and increased expression of matrix metalloproteinase 16 via miR 200b downregulation in bladder cancer cells. Mol Med Rep. 2014; 10:1549–1554.
40. Majid S, Dar AA, Saini S, Deng G, Chang I, Greene K, et al. MicroRNA-23b functions as a tumor suppressor by regulating Zeb1 in bladder cancer. PLoS One. 2013; 8:e67686.
41. Sun DK, Wang JM, Zhang P, Wang YQ. MicroRNA-138 regulates metastatic potential of bladder cancer through ZEB2. Cell Physiol Biochem. 2015; 37:2366–2374.
42. Kenney PA, Wszolek MF, Rieger-Christ KM, Neto BS, Gould JJ, Harty NJ, et al. Novel ZEB1 expression in bladder tumorigenesis. BJU Int. 2011; 107:656–663.
43. Zhou Y, Wu D, Tao J, Qu P, Zhou Z, Hou J. MicroRNA-133 inhibits cell proliferation, migration and invasion by targeting epidermal growth factor receptor and its downstream effector proteins in bladder cancer. Scand J Urol. 2013; 47:423–432.
44. Zhu Z, Xu T, Wang L, Wang X, Zhong S, Xu C, et al. MicroRNA-145 directly targets the insulin-like growth factor receptor I in human bladder cancer cells. FEBS Lett. 2014; 588:3180–3185.
45. Gomez-Roman JJ, Saenz P, Molina M, Cuevas Gonzalez J, Escuredo K, Santa Cruz S, et al. Fibroblast growth factor receptor 3 is overexpressed in urinary tract carcinomas and modulates the neoplastic cell growth. Clin Cancer Res. 2005; 11(2 Pt 1):459–465.
46. Catto JW, Miah S, Owen HC, Bryant H, Myers K, Dudziec E, et al. Distinct microRNA alterations characterize high- and low-grade bladder cancer. Cancer Res. 2009; 69:8472–8481.
47. Chen XN, Wang KF, Xu ZQ, Li SJ, Liu Q, Fu DH, et al. MiR-133b regulates bladder cancer cell proliferation and apoptosis by targeting Bcl-w and Akt1. Cancer Cell Int. 2014; 14:70.
48. Noguchi S, Yasui Y, Iwasaki J, Kumazaki M, Yamada N, Naito S, et al. Replacement treatment with microRNA-143 and -145 induces synergistic inhibition of the growth of human bladder cancer cells by regulating PI3K/Akt and MAPK signaling pathways. Cancer Lett. 2013; 328:353–361.
49. Zhou XU, Qi L, Tong S, Cui YU, Chen J, Huang T, et al. miR-128 downregulation promotes growth and metastasis of bladder cancer cells and involves VEGF-C upregulation. Oncol Lett. 2015; 10:3183–3190.
50. Ueno K, Hirata H, Majid S, Yamamura S, Shahryari V, Tabatabai ZL, et al. Tumor suppressor microRNA-493 decreases cell motility and migration ability in human bladder cancer cells by downregulating RhoC and FZD4. Mol Cancer Ther. 2012; 11:244–253.
51. Deng H, Lv L, Li Y, Zhang C, Meng F, Pu Y, et al. The miR-193a-3p regulated PSEN1 gene suppresses the multi-chemoresistance of bladder cancer. Biochim Biophys Acta. 2015; 1852:520–528.
52. Lei Y, Li B, Tong S, Qi L, Hu X, Cui Y, et al. miR-101 suppresses vascular endothelial growth factor C that inhibits migration and invasion and enhances cisplatin chemosensitivity of bladder cancer cells. PLoS One. 2015; 10:e0117809.
53. Hanke M, Hoefig K, Merz H, Feller AC, Kausch I, Jocham D, et al. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol Oncol. 2010; 28:655–661.
54. Yamada Y, Enokida H, Kojima S, Kawakami K, Chiyomaru T, Tatarano S, et al. MiR-96 and miR-183 detection in urine serve as potential tumor markers of urothelial carcinoma: correlation with stage and grade, and comparison with urinary cytology. Cancer Sci. 2011; 102:522–529.
55. Shimizu T, Suzuki H, Nojima M, Kitamura H, Yamamoto E, Maruyama R, et al. Methylation of a panel of microRNA genes is a novel biomarker for detection of bladder cancer. Eur Urol. 2013; 63:1091–1100.
56. Long JD, Sullivan TB, Humphrey J, Logvinenko T, Summerhayes KA, Kozinn S, et al. A non-invasive miRNA based assay to detect bladder cancer in cell-free urine. Am J Transl Res. 2015; 7:2500–2509.
57. Wang J, Zhang X, Wang L, Dong Z, Du L, Yang Y, et al. Downregulation of urinary cell-free microRNA-214 as a diagnostic and prognostic biomarker in bladder cancer. J Surg Oncol. 2015; 111:992–999.
58. Zhang DZ, Lau KM, Chan ES, Wang G, Szeto CC, Wong K, et al. Cell-free urinary microRNA-99a and microRNA-125b are diagnostic markers for the non-invasive screening of bladder cancer. PLoS One. 2014; 9:e100793.
59. Zhou X, Zhang X, Yang Y, Li Z, Du L, Dong Z, et al. Urinary cell-free microRNA-106b as a novel biomarker for detection of bladder cancer. Med Oncol. 2014; 31:197.
60. Yun SJ, Jeong P, Kim WT, Kim TH, Lee YS, Song PH, et al. Cell-free microRNAs in urine as diagnostic and prognostic biomarkers of bladder cancer. Int J Oncol. 2012; 41:1871–1878.
61. Mengual L, Lozano JJ, Ingelmo-Torres M, Gazquez C, Ribal MJ, Alcaraz A. Using microRNA profiling in urine samples to develop a non-invasive test for bladder cancer. Int J Cancer. 2013; 133:2631–2641.
62. Dudziec E, Miah S, Choudhry HM, Owen HC, Blizard S, Glover M, et al. Hypermethylation of CpG islands and shores around specific microRNAs and mirtrons is associated with the phenotype and presence of bladder cancer. Clin Cancer Res. 2011; 17:1287–1296.
63. Jiang X, Du L, Wang L, Li J, Liu Y, Zheng G, et al. Serum microRNA expression signatures identified from genome-wide microRNA profiling serve as novel noninvasive biomarkers for diagnosis and recurrence of bladder cancer. Int J Cancer. 2015; 136:854–862.
64. Kim SM, Kang HW, Kim WT, Kim YJ, Yun SJ, Lee SC, et al. Cell-free microRNA-214 from urine as a biomarker for non-muscle-invasive bladder cancer. Korean J Urol. 2013; 54:791–796.
65. Zhang X, Zhang Y, Liu X, Fang A, Li P, Li Z, et al. MicroRNA-203 is a prognostic indicator in bladder cancer and enhances chemosensitivity to cisplatin via apoptosis by targeting Bcl-w and survivin. PLoS One. 2015; 10:e0143441.
66. Yu G, Yao W, Xiao W, Li H, Xu H, Lang B. MicroRNA-34a functions as an anti-metastatic microRNA and suppresses angiogenesis in bladder cancer by directly targeting CD44. J Exp Clin Cancer Res. 2014; 33:779.
67. Cheng Y, Yang X, Deng X, Zhang X, Li P, Tao J, et al. MicroRNA-218 inhibits bladder cancer cell proliferation, migration, and invasion by targeting BMI-1. Tumour Biol. 2015; 36:8015–8023.
68. Xu C, Zeng Q, Xu W, Jiao L, Chen Y, Zhang Z, et al. miRNA-100 inhibits human bladder urothelial carcinogenesis by directly targeting mTOR. Mol Cancer Ther. 2013; 12:207–219.
69. Peng Y, Dong W, Lin TX, Zhong GZ, Liao B, Wang B, et al. MicroRNA-155 promotes bladder cancer growth by repressing the tumor suppressor DMTF1. Oncotarget. 2015; 6:16043–16058.
70. Xiao H, Li H, Yu G, Xiao W, Hu J, Tang K, et al. MicroRNA-10b promotes migration and invasion through KLF4 and HOXD10 in human bladder cancer. Oncol Rep. 2014; 31:1832–1838.
71. Inamoto T, Taniguchi K, Takahara K, Iwatsuki A, Takai T, Komura K, et al. Intravesical administration of exogenous microRNA-145 as a therapy for mouse orthotopic human bladder cancer xenograft. Oncotarget. 2015; 6:21628–21635.
72. Liu Y, Han Y, Zhang H, Nie L, Jiang Z, Fa P, et al. Synthetic miRNA-mowers targeting miR-183-96-182 cluster or miR-210 inhibit growth and migration and induce apoptosis in bladder cancer cells. PLoS One. 2012; 7:e52280.
73. Yamasaki T, Yoshino H, Enokida H, Hidaka H, Chiyomaru T, Nohata N, et al. Novel molecular targets regulated by tumor suppressors microRNA-1 and microRNA-133a in bladder cancer. Int J Oncol. 2012; 40:1821–1830.
74. Yoshino H, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Nishiyama K, et al. The tumour-suppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. Br J Cancer. 2011; 104:808–818.
75. Jiang QQ, Liu B, Yuan T. MicroRNA-16 inhibits bladder cancer proliferation by targeting Cyclin D1. Asian Pac J Cancer Prev. 2013; 14:4127–4130.
76. Zhang S, Zhang C, Liu W, Zheng W, Zhang Y, Wang S, et al. MicroRNA-24 upregulation inhibits proliferation, metastasis and induces apoptosis in bladder cancer cells by targeting CARMA3. Int J Oncol. 2015; 47:1351–1360.
77. Deng Y, Bai H, Hu H. rs11671784 G/A variation in miR-27a decreases chemo-sensitivity of bladder cancer by decreasing miR-27a and increasing the target RUNX-1 expression. Biochem Biophys Res Commun. 2015; 458:321–327.
78. Drayton RM, Dudziec E, Peter S, Bertz S, Hartmann A, Bryant HE, et al. Reduced expression of miRNA-27a modulates cisplatin resistance in bladder cancer by targeting the cystine/glutamate exchanger SLC7A11. Clin Cancer Res. 2014; 20:1990–2000.
79. Yuan L, Chu H, Wang M, Gu X, Shi D, Ma L, et al. Genetic variation in DROSHA 3'UTR regulated by hsa-miR-27b is associated with bladder cancer risk. PLoS One. 2013; 8:e81524.
80. Zhao X, Li J, Huang S, Wan X, Luo H, Wu D. MiRNA-29c regulates cell growth and invasion by targeting CDK6 in bladder cancer. Am J Transl Res. 2015; 7:1382–1389.
81. Zhang C, Ma X, Du J, Yao Z, Shi T, Ai Q, et al. MicroRNA-30a as a prognostic factor in Urothelial Carcinoma of Bladder inhibits cellular malignancy by antagonizing Notch1. BJU Int. 2016; 01. 18. [Epub]. DOI: 10.1111/bju.13407.
82. Sun H, Tian J, Xian W, Xie T, Yang X. miR-34a inhibits proliferation and invasion of bladder cancer cells by targeting orphan nuclear receptor HNF4G. Dis Markers. 2015; 2015:879254.
83. Zhang C, Yao Z, Zhu M, Ma X, Shi T, Li H, et al. Inhibitory effects of microRNA-34a on cell migration and invasion of invasive urothelial bladder carcinoma by targeting Notch1. J Huazhong Univ Sci Technolog Med Sci. 2012; 32:375–382.
84. Bu Q, Fang Y, Cao Y, Chen Q, Liu Y. Enforced expression of miR-101 enhances cisplatin sensitivity in human bladder cancer cells by modulating the cyclooxygenase-2 pathway. Mol Med Rep. 2014; 10:2203–2209.
85. Hu Z, Lin Y, Chen H, Mao Y, Wu J, Zhu Y, et al. MicroRNA-101 suppresses motility of bladder cancer cells by targeting c-Met. Biochem Biophys Res Commun. 2013; 435:82–87.
86. Xu X, Li S, Lin Y, Chen H, Hu Z, Mao Y, et al. MicroRNA-124-3p inhibits cell migration and invasion in bladder cancer cells by targeting ROCK1. J Transl Med. 2013; 11:276.
87. Zhang T, Wang J, Zhai X, Li H, Li C, Chang J. MiR-124 retards bladder cancer growth by directly targeting CDK4. Acta Biochim Biophys Sin (Shanghai). 2014; 46:1072–1079.
88. Wu D, Ding J, Wang L, Pan H, Zhou Z, Zhou J, et al. microRNA-125b inhibits cell migration and invasion by targeting matrix metallopeptidase 13 in bladder cancer. Oncol Lett. 2013; 5:829–834.
89. Zhao X, He W, Li J, Huang S, Wan X, Luo H, et al. MiRNA-125b inhibits proliferation and migration by targeting SphK1 in bladder cancer. Am J Transl Res. 2015; 7:2346–2354.
90. Uchida Y, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Kawahara K, et al. MiR-133a induces apoptosis through direct regulation of GSTP1 in bladder cancer cell lines. Urol Oncol. 2013; 31:115–123.
91. Mao XP, Zhang LS, Huang B, Zhou SY, Liao J, Chen LW, et al. Mir-135a enhances cellular proliferation through post-transcriptionally regulating PHLPP2 and FOXO1 in human bladder cancer. J Transl Med. 2015; 13:86.
92. Ostenfeld MS, Bramsen JB, Lamy P, Villadsen SB, Fristrup N, Sorensen KD, et al. miR-145 induces caspase-dependent and -independent cell death in urothelial cancer cell lines with targeting of an expression signature present in Ta bladder tumors. Oncogene. 2010; 29:1073–1084.
93. Kou B, Gao Y, Du C, Shi Q, Xu S, Wang CQ, et al. miR-145 inhibits invasion of bladder cancer cells by targeting PAK1. Urol Oncol. 2014; 32:846–854.
94. Noguchi S, Yamada N, Kumazaki M, Yasui Y, Iwasaki J, Naito S, et al. socs7, a target gene of microRNA-145, regulates interferon-β induction through STAT3 nuclear translocation in bladder cancer cells. Cell Death Dis. 2013; 4:e482.
95. Yao K, He L, Gan Y, Zeng Q, Dai Y, Tan J. MiR-186 suppresses the growth and metastasis of bladder cancer by targeting NSBP1. Diagn Pathol. 2015; 10:146.
96. Deng H, Lv L, Li Y, Zhang C, Meng F, Pu Y, et al. miR-193a-3p regulates the multi-drug resistance of bladder cancer by targeting the LOXL4 gene and the oxidative stress pathway. Mol Cancer. 2014; 13:234.
97. Lv L, Li Y, Deng H, Zhang C, Pu Y, Qian L, et al. MiR-193a-3p promotes the multi-chemoresistance of bladder cancer by targeting the HOXC9 gene. Cancer Lett. 2015; 357:105–113.
98. Zhao C, Qi L, Chen M, Liu L, Yan W, Tong S, et al. microRNA-195 inhibits cell proliferation in bladder cancer via inhibition of cell division control protein 42 homolog/signal transducer and activator of transcription-3 signaling. Exp Ther Med. 2015; 10:1103–1108.
99. Lin Y, Wu J, Chen H, Mao Y, Liu Y, Mao Q, et al. Cyclin-dependent kinase 4 is a novel target in micoRNA-195-mediated cell cycle arrest in bladder cancer cells. FEBS Lett. 2012; 586:442–447.
100. Fei X, Qi M, Wu B, Song Y, Wang Y, Li T. MicroRNA-195-5p suppresses glucose uptake and proliferation of human bladder cancer T24 cells by regulating GLUT3 expression. FEBS Lett. 2012; 586:392–397.
101. Wang J, Zhang X, Wang L, Yang Y, Dong Z, Wang H, et al. MicroRNA-214 suppresses oncogenesis and exerts impact on prognosis by targeting PDRG1 in bladder cancer. PLoS One. 2015; 10:e0118086.
102. Liu J, Cao J, Zhao X. miR-221 facilitates the TGFbeta1-induced epithelial-mesenchymal transition in human bladder cancer cells by targeting STMN1. BMC Urol. 2015; 15:36.
103. Shang C, Zhang H, Guo Y, Hong Y, Liu Y, Xue Y. MiR-320a down-regulation mediates bladder carcinoma invasion by targeting ITGB3. Mol Biol Rep. 2014; 41:2521–2527.
104. Wang X, Wu J, Lin Y, Zhu Y, Xu X, Xu X, et al. MicroRNA-320c inhibits tumorous behaviors of bladder cancer by targeting Cyclin-dependent kinase 6. J Exp Clin Cancer Res. 2014; 33:69.
105. Chen H, Lin YW, Mao YQ, Wu J, Liu YF, Zheng XY, et al. MicroRNA-449a acts as a tumor suppressor in human bladder cancer through the regulation of pocket proteins. Cancer Lett. 2012; 320:40–47.
106. Chen Z, Li Q, Wang S, Zhang J. miR 485 5p inhibits bladder cancer metastasis by targeting HMGA2. Int J Mol Med. 2015; 36:1136–1142.
107. Li S, Xu X, Xu X, Hu Z, Wu J, Zhu Y, et al. MicroRNA-490-5p inhibits proliferation of bladder cancer by targeting c-Fos. Biochem Biophys Res Commun. 2013; 441:976–981.
108. Tatarano S, Chiyomaru T, Kawakami K, Enokida H, Yoshino H, Hidaka H, et al. Novel oncogenic function of mesoderm development candidate 1 and its regulation by MiR-574-3p in bladder cancer cell lines. Int J Oncol. 2012; 40:951–959.
109. Liang Z, Li S, Xu X, Xu X, Wang X, Wu J, et al. MicroRNA-576-3p inhibits proliferation in bladder cancer cells by targeting cyclin D1. Mol Cells. 2015; 38:130–137.
110. Mo M, Peng F, Wang L, Peng L, Lan G, Yu S. Roles of mitochondrial transcription factor A and microRNA-590-3p in the development of bladder cancer. Oncol Lett. 2013; 6:617–623.
111. Zhou J, Dai W, Song J. miR-1182 inhibits growth and mediates the chemosensitivity of bladder cancer by targeting hTERT. Biochem Biophys Res Commun. 2016; 470:445–452.
112. Xie D, Shang C, Zhang H, Guo Y, Tong X. Up-regulation of miR-9 target CBX7 to regulate invasion ability of bladder transitional cell carcinoma. Med Sci Monit. 2015; 21:225–230.
113. Wang H, Zhang W, Zuo Y, Ding M, Ke C, Yan R, et al. miR-9 promotes cell proliferation and inhibits apoptosis by targeting LASS2 in bladder cancer. Tumour Biol. 2015; 36:9631–9640.
114. Cao Y, Yu SL, Wang Y, Guo GY, Ding Q, An RH. MicroRNA-dependent regulation of PTEN after arsenic trioxide treatment in bladder cancer cell line T24. Tumour Biol. 2011; 32:179–188.
115. Lei Y, Hu X, Li B, Peng M, Tong S, Zu X, et al. miR-150 modulates cisplatin chemosensitivity and invasiveness of muscle-invasive bladder cancer cells via targeting PDCD4 in vitro. Med Sci Monit. 2014; 20:1850–1857.
116. Hirata H, Ueno K, Shahryari V, Tanaka Y, Tabatabai ZL, Hinoda Y, et al. Oncogenic miRNA-182-5p targets Smad4 and RECK in human bladder cancer. PLoS One. 2012; 7:e51056.
117. Tolle A, Jung M, Rabenhorst S, Kilic E, Jung K, Weikert S. Identification of microRNAs in blood and urine as tumour markers for the detection of urinary bladder cancer. Oncol Rep. 2013; 30:1949–1956.
118. Miah S, Dudziec E, Drayton RM, Zlotta AR, Morgan SL, Rosario DJ, et al. An evaluation of urinary microRNA reveals a high sensitivity for bladder cancer. Br J Cancer. 2012; 107:123–128.
119. Motawi TK, Rizk SM, Ibrahim TM, Ibrahim IA. Circulating microRNAs, miR-92a, miR-100 and miR-143, as non-invasive biomarkers for bladder cancer diagnosis. Cell Biochem Funct. 2016; 34:142–148.
120. Fang Z, Dai W, Wang X, Chen W, Shen C, Ye G, et al. Circulating miR-205: a promising biomarker for the detection and prognosis evaluation of bladder cancer. Tumour Biol. 2015; 12. 29. [Epub]. DOI: 10.1007/s13277-015-4698-y.
121. Du M, Shi D, Yuan L, Li P, Chu H, Qin C, et al. Circulating miR-497 and miR-663b in plasma are potential novel biomarkers for bladder cancer. Sci Rep. 2015; 5:10437.
122. Zhang HH, Qi F, Cao YH, Zu XB, Chen MF. Expression and clinical significance of microRNA-21, maspin and vascular endothelial growth factor-C in bladder cancer. Oncol Lett. 2015; 10:2610–2616.
123. Wu CT, Lin WY, Chang YH, Lin PY, Chen WC, Chen MF. DNMT1-dependent suppression of microRNA424 regulates tumor progression in human bladder cancer. Oncotarget. 2015; 6:24119–24131.
124. Xu Z, Yu YQ, Ge YZ, Zhu JG, Zhu M, Zhao YC, et al. MicroRNA expression profiles in muscle-invasive bladder cancer: identification of a four-microRNA signature associated with patient survival. Tumour Biol. 2015; 36:8159–8166.
125. Avgeris M, Mavridis K, Tokas T, Stravodimos K, Fragoulis EG, Scorilas A. Uncovering the clinical utility of miR-143, miR-145 and miR-224 for predicting the survival of bladder cancer patients following treatment. Carcinogenesis. 2015; 36:528–537.
126. Lin R, Shen W, Zhi Y, Zhou Z. Prognostic value of miR-26a and HMGA1 in urothelial bladder cancer. Biomed Pharmacother. 2014; 68:929–934.
127. Wang XL, Xie HY, Zhu CD, Zhu XF, Cao GX, Chen XH, et al. Increased miR-141 expression is associated with diagnosis and favorable prognosis of patients with bladder cancer. Tumour Biol. 2015; 36:877–883.
128. Zhang DQ, Zhou CK, Jiang XW, Chen J, Shi BK. Increased expression of miR-222 is associated with poor prognosis in bladder cancer. World J Surg Oncol. 2014; 12:241.
129. Segersten U, Spector Y, Goren Y, Tabak S, Malmström PU. The role of microRNA profiling in prognosticating progression in Ta and T1 urinary bladder cancer. Urol Oncol. 2014; 32:613–618.
130. Ratert N, Meyer HA, Jung M, Lioudmer P, Mollenkopf HJ, Wagner I, et al. miRNA profiling identifies candidate mirnas for bladder cancer diagnosis and clinical outcome. J Mol Diagn. 2013; 15:695–705.
131. Wang S, Li Q, Wang K, Dai Y, Yang J, Xue S, et al. Decreased expression of microRNA-31 associates with aggressive tumor progression and poor prognosis in patients with bladder cancer. Clin Transl Oncol. 2013; 15:849–854.
132. Rosenberg E, Baniel J, Spector Y, Faerman A, Meiri E, Aharonov R, et al. Predicting progression of bladder urothelial carcinoma using microRNA expression. BJU Int. 2013; 112:1027–1034.
133. Wang S, Xue S, Dai Y, Yang J, Chen Z, Fang X, et al. Reduced expression of microRNA-100 confers unfavorable prognosis in patients with bladder cancer. Diagn Pathol. 2012; 7:159.
134. Pignot G, Cizeron-Clairac G, Vacher S, Susini A, Tozlu S, Vieillefond A, et al. microRNA expression profile in a large series of bladder tumors: identification of a 3-miRNA signature associated with aggressiveness of muscle-invasive bladder cancer. Int J Cancer. 2013; 132:2479–2491.
135. Majid S, Dar AA, Saini S, Shahryari V, Arora S, Zaman MS, et al. MicroRNA-1280 inhibits invasion and metastasis by targeting ROCK1 in bladder cancer. PLoS One. 2012; 7:e46743.
136. Zaravinos A, Radojicic J, Lambrou GI, Volanis D, Delakas D, Stathopoulos EN, et al. Expression of miRNAs involved in angiogenesis, tumor cell proliferation, tumor suppressor inhibition, epithelial-mesenchymal transition and activation of metastasis in bladder cancer. J Urol. 2012; 188:615–623.
137. Puerta-Gil P, Garcia-Baquero R, Jia AY, Ocana S, Alvarez-Mugica M, Alvarez-Ossorio JL, et al. miR-143, miR-222, and miR-452 are useful as tumor stratification and noninvasive diagnostic biomarkers for bladder cancer. Am J Pathol. 2012; 180:1808–1815.
138. Pignot G, Vieillefond A, Vacher S, Zerbib M, Debre B, Lidereau R, et al. Hedgehog pathway activation in human transitional cell carcinoma of the bladder. Br J Cancer. 2012; 106:1177–1186.
139. Liu W, Qi L, Lv H, Zu X, Chen M, Wang J, et al. MiRNA-141 and miRNA-200b are closely related to invasive ability and considered as decision-making biomarkers for the extent of PLND during cystectomy. BMC Cancer. 2015; 15:92.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download