Abstract
Serum or plasma is free of cellular components. As DNA is in the nucleus or mitochondria of a cell, it can be presumed that serum/plasma is DNA free. However, there are cases wherein serum/plasma is the only resource available for identification analysis, yet no sufficient data are available regarding whether reliable DNA testing can be applied to such cases, and what the influencing factors are when testing is a valid course of action. The aim of this study is to illustrate the factors that can be used in the genetic testing of serum/plasma when identifying an individual. The results showed that the concentration of serum DNA significantly increased over time in 4°C storage, and the DNA yields from samples stored in heparin tubes were overall higher than from samples stored in ethylenediaminetetraacetic acid tubes. We observed that the concentration of DNA in serum successfully matched 100% to the short tandem repeat data of blood DNA.
REFERENCES
1.Lo YM., Tein MS., Lau TK, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998. 62:768–75.

2.Lee JH., Lee HY., Cho S, et al. DNA profiling via short tandem repeat analysis by using serum samples. Korean J Leg Med. 2013. 37:220–3.

3.Takayama T., Yamada S., Watanabe Y, et al. Origin of DNA in human serum and usefulness of serum as a material for DNA typing. Leg Med (Tokyo). 2001. 3:109–13.

4.Honda H., Miharu N., Ohashi Y, et al. Fetal gender determination in early pregnancy through qualitative and quantitative analysis of fetal DNA in maternal serum. Hum Genet. 2002. 110:75–9.

5.Bischoff FZ., Sinacori MK., Dang DD, et al. Cell-free fetal DNA and intact fetal cells in maternal blood circulation: implications for first and second trimester non-invasive prenatal diagnosis. Hum Reprod Update. 2002. 8:493–500.

6.Allen RW., Pritchard JK. Resolution of a serum sample mix-up through the use of short tandem repeat DNA typing. Transfusion. 2004. 44:1750–4.

7.Lam NY., Rainer TH., Chiu RW, et al. EDTA is a better anticoagulant than heparin or citrate for delayed blood processing for plasma DNA analysis. Clin Chem. 2004. 50:256–7.

8.Ravard-Goulvestre C., Crainic K., Guillon F, et al. Successful extraction of human genomic DNA from serum and its application to forensic identification. J Forensic Sci. 2004. 49:60–3.

Fig. 1.
Box plots of DNA concentrations in serum (A, B) and plasma (C). RT, room temperature; EDTA, ethylenediaminetetraacetic acid.
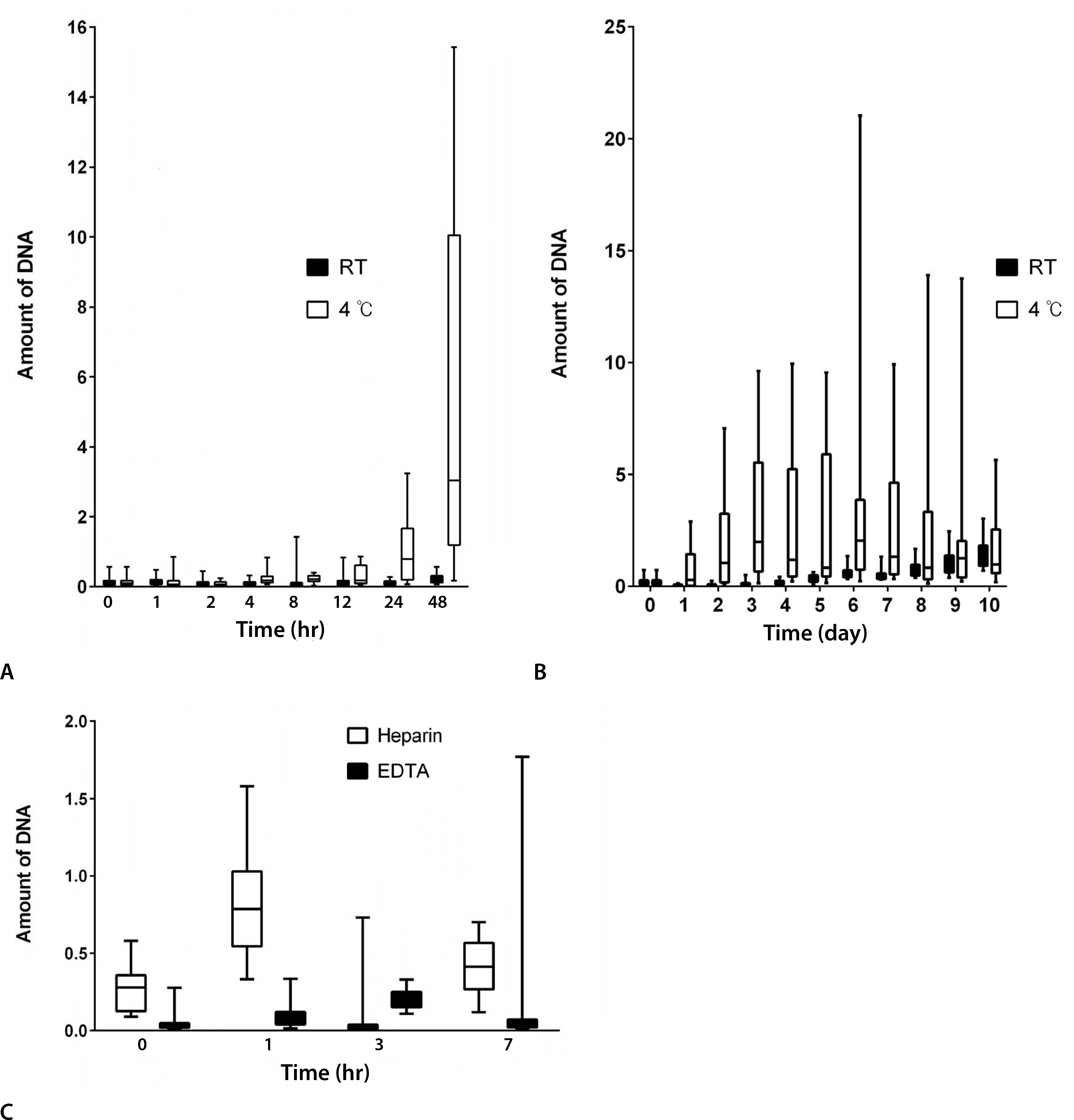
Fig. 2.
Electropherograms of the amplified DNA from one sample. (A) Peak exceeding the 50-RFU threshold is present. (B) Peaks exceeding the 40-RFU threshold are present. RFU, relative fluorescence unit.
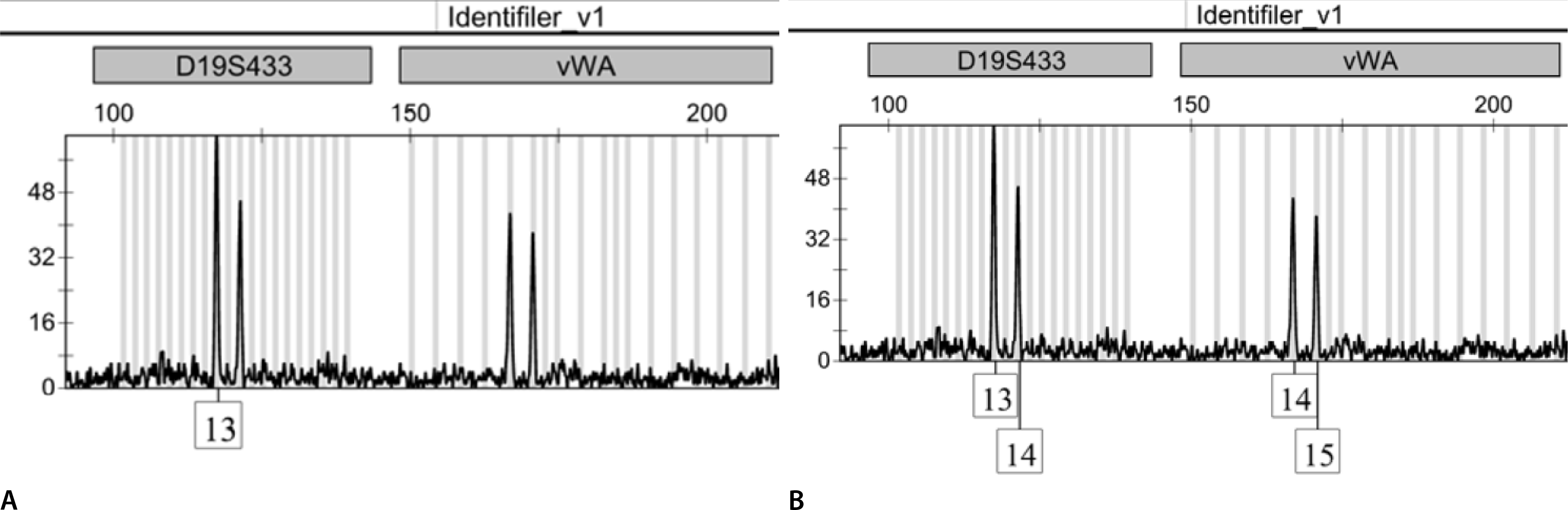
Table 1.
Amount of DNA recovered from donors (15 and 14) with time-temperature differences by two-way ANOVA
Table 2.
Amount of DNA recovered from 15 donors with time-anticoagulant (heparin, EDTA) differences by two-way ANOVA




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download