Abstract
Diabetes is a common metabolic disorder with a worldwide prevalence of 8.3% and is the leading cause of visual loss, end-stage renal disease and amputation. Recently, genome-wide association studies (GWASs) have identified genetic risk factors for diabetic microvascular complications of retinopathy, nephropathy, and neuropathy. We summarized the recent findings of GWASs on diabetic microvascular complications and highlighted the challenges and our opinion on future directives. Five GWASs were conducted on diabetic retinopathy, nine on nephropathy, and one on neuropathic pain. The majority of recent GWASs were underpowered and heterogeneous in terms of study design, inclusion criteria and phenotype definition. Therefore, few reached the genome-wide significance threshold and the findings were inconsistent across the studies. Recent GWASs provided novel information on genetic risk factors and the possible pathophysiology of diabetic microvascular complications. However, further collaborative efforts to standardize phenotype definition and increase sample size are necessary for successful genetic studies on diabetic microvascular complications.
Diabetes mellitus is a chronic metabolic disorder that can result in multiple long-term micro- and macrovascular complications. Microvascular complications include retinopathy, nephropathy and neuropathy. Diabetes is well known as the leading cause of blindness, end-stage renal disease (ESRD) and limb amputation. In Korea, approximately 18.6% of diabetic patients have retinopathy, 27.3% albuminuria, and 33.5% diabetic neuropathy [1]. In a nationwide survey in 2012, diabetes accounted for 50.6% of new-onset ESRD in Korea [2]. Microvascular complications significantly affect the quality of life and impose a major burden on the healthcare system and economy.
The development of microvascular complications is related to several environmental risk factors, including duration of diabetes, degree of hyperglycemia, blood pressure, and dyslipidemia. In the landmark U.K. Prospective Diabetes Study, which enrolled newly diagnosed type 2 diabetes mellitus (T2DM) patients, participants randomized to intensive glucose control (median hemoglobin A1c [HbA1c] 7.0%) had a 25% reduction in microvascular complications including vitreous hemorrhage, retinal photocoagulation and ESRD compared with those in the conventional treatment group (median HbA1c 7.9%) after 10 years of follow-up [3]. In the recent Action to Control Cardiovascular Risk in Diabetes (ACCORD) trial, intensive glycemic control targeting HbA1c <6.0% resulted in 23% reduction of retinopathy progression [4] and delayed onset of albuminuria and peripheral neuropathy [5]. Treatment with fenofibrate also decreased the risk of retinopathy in the ACCORD Eye Study [4].
Despite various interventions to control these environmental factors, large individual variations in the outcome of diabetic microvascular complications exist. Some patients with a short duration of diabetes develop microvascular complications although they had relatively good glycemic control. In contrast, some people do not develop microvascular complications even with a prolonged disease duration and with poor glycemic control. These clinical findings suggest that genetic factors play a role in the pathogenesis of microvascular complications. Familial aggregation in diabetic microvascular complications provides further evidence of genetic predisposition. For example, the heritability for diabetic retinopathy was estimated at 18% [6] and for proliferative diabetic retinopathy (PDR) at 52% [7]. The development of diabetic nephropathy also differs based on ethnicity and African Americans and Asians have 1.9- and 1.8-fold increased risks of ESRD, respectively, compared with European diabetic patients [8]. Consequently, efforts have been made to identify genetic risk factors for diabetic microvascular complications using candidate gene approach, linkage analysis and the recent genome-wide association studies (GWASs).
Based on biological or positional plausibility, numerous candidate genes were selected for genetic association studies. The most thoroughly investigated genes include vascular endothelial growth factor A (VEGFA) [9], aldo-keto reductase family 1, member B1 (AKR1B1) [10], and erythropoietin (EPO) [11] for diabetic retinopathy and angiotensin 1 converting enzyme (ACE) [12], protein kinase C β (PRKCB) [13], and erythropoietin (EPO) [11] for diabetic nephropathy. Although significant associations were initially reported, subsequent studies frequently showed inconsistent results. Most of the candidate gene association studies were conducted using a small sample size and had less stringent statistical thresholds. In addition, a meta-analysis showed that these candidate variants were not significantly associated on a genome-wide basis [14].
The advances in genotyping technology and publicly available databases of reference genomes and human genetic variations, including the International HapMap project [15], have contributed to understanding the genetic risk factors of common metabolic disorders using GWASs. In GWASs, hundreds of thousands or more of single-nucleotide variants are genotyped and tested for association with a disease or a continuous trait in several hundred or more subjects [16]. GWASs do not rely on previous knowledge and thus, are free from bias. Recently, GWASs have increased the number of genetic markers to more than one million by imputation methods and the sample size has increased to more than one hundred thousand by using meta-GWASs. The first successful GWAS on T2DM was published in 2007 [17]. Since then, at least 77 confirmed genetic loci for T2DM have been identified, [18] providing a better understanding of diabetes pathophysiology and there are ongoing efforts to use this genetic information in risk prediction and tailoring of individualized therapy [19]. In parallel, attempts have been made to unravel the genetic risk factors for diabetic microvascular complications using GWASs. In this article, we reviewed the recent GWASs on diabetic retinopathy, nephropathy and neuropathy and discussed their limitations and future directives.
Diabetic retinopathy is clinically defined by the retinal microvascular lesions in diabetic patients and broadly classified into nonproliferative diabetic retinopathy (NPDR) and PDR. Diabetic macular edema can result in moderate visual loss and can be present at any stage of diabetic retinopathy, although more common in advanced retinopathy. The gold standard for classification of diabetic retinopathy severity is derived from the Early Treatment of Diabetic Retinopathy Study [20]. Diabetic retinopathy increases as the duration of diabetes increases. According to the Wisconsin Epidemiologic Study of Diabetic Retinopathy in T2DM patients, diabetic retinopathy is present in 20% of patients at the time of diagnosis, which increases to 60% to 85% after 15 years [21]. Whether the pathophysiology of retinopathy differs between type 1 diabetes mellitus (T1DM) and T2DM remains unknown. All heterogeneity factors-including the severity, duration, and type of diabetes-should be considered to understand the genetic risk factors for diabetic retinopathy.
Currently, five GWASs on diabetic retinopathy have been published. The first GWAS performed included 283 Mexican-American T2DM retinopathy patients and controls (Table 1) [2223242526]. Fu et al. [22] found two potential loci in the introns of calcium/calmodulin-dependent protein kinase IV (CAMK4) and formin 1 (FMN1) genes. Huang et al. [23] reported a GWAS on T2DM diabetic retinopathy including 749 Taiwanese patients and found seven independent loci with potential significance (P<1.0×10-6). However, they reported the lowest P value among the six genetic models (genotype, allele, trend, additive, dominant, and recessive) and did not adjust for multiple comparisons. The third study is the largest GWAS conducted to date and is a meta-analysis of two GWASs, Genetics of Kidneys in Diabetes (GoKinD) and Epidemiology of Diabetes Interventions and Complications (EDIC) studies [24]. This study by Grassi et al. [24] involved 2,829 European subjects with T1DM. The most significant variant was rs476141 located in a long non-coding RNA (LOC339529) in chromosome 1 with P values of 1.20×10-7. The study by Sheu et al. [25] used a two-stage GWAS with follow-up genotyping in an independent population. Although they found three potential genetic variants in stage 1 GWAS in Taiwanese subjects, these findings were not replicated in the Hispanic population. The latest GWAS on diabetic retinopathy was a three-stage design performed by Awata et al. [26] in Japanese subjects. Among the eight variants that were followed-up to the third stage, none reached a genome-wide significance threshold. The most significant variant was located in RP1-90L14.1, a long non-coding RNA gene, with a P value of 1.70×10-5 in the meta-analysis of the three-stage results.
Overall, three studies were performed with Asian subjects, one with Mexican-American and one with European subjects. One study included T1DM patients and the remaining four studies included T2DM patients. The earliest two studies performed by Fu et al. [22] and Huang et al. [23] were single-stage GWASs with a small sample of fewer than 1,000 subjects. The findings were not replicated in an independent cohort using a different genotyping method. The majority of the genetic variants reported from the five studies did not pass the conventional significance threshold of P<5.0×10-8, except for several variants in the study by Huang et al. [23]. However, the latter study reported the best P value among various genetic models and did not correct for multiple comparisons. None of the genetic variants reported overlapped among the five studies; however, cases and controls were defined differently, which could be a crucial point when performing a genetic study. Regarding case groups, several studies included subjects with either NPDR or PDR, whereas others included only subjects with PDR. Regarding the control group, only in the study by Sheu et al. [25] the subjects were limited to those with a diabetes duration of more than 8 years without any diabetic retinopathy. The heterogeneity in study design and relatively small sample sizes could explain the inconsistencies in the genetic variants identified in the five GWASs on diabetic microvascular complications. A clear and rational definition of cases and controls is necessary to enhance genetic contrast as well as a large sample size to ensure sufficient statistical power.
Diabetic nephropathy is clinically defined as an increase in urinary albumin excretion and a decrease in kidney function. Classification of diabetic nephropathy using the Kidney Disease: Improving Global Outcomes group criteria is based on estimated glomerular filtration rate (eGFR) and the degree of proteinuria. The eGFR is generally calculated using the Modification of Diet in Renal Disease formula and is divided into five stages. The degree of proteinuria is used for substaging diabetic nephropathy. The eGFR reflects the current kidney function and proteinuria reflects the extent of pathological kidney damage. Proteinuria is a hallmark of diabetic nephropathy and precedes the decline in kidney function, but is not a prerequisite in some cases. A large body of evidence indicates that treatments to prevent or delay its progression should include intensive glycemic and blood pressure control. In the Action in Diabetes and Vascular Disease (ADVANCE) trial, intensive glycemic control resulted in the risk reduction for microalbuminuria (30 to 300 mg/g), macroalbuminuria (>300 mg/g) and ESRD, by 9%, 30%, and 65%, respectively [27]. In the pivotal study of Diabetes Control and Complications Trial (DCCT), intensive glucose control in T1DM patients resulted in 39% reduced occurrence of microalbuminuria [28]. However, 25% of participants in the intensive treatment group eventually developed microalbuminuria during the 6.5-year follow-up period. Therefore, individual variations in the risk of diabetic nephropathy exist and genetic factors likely play an important role.
Consequently, efforts have been made to understand the genetic risk factors for diabetic nephropathy. Nine GWASs on diabetic nephropathy have been published (Table 2) [293031323334353637]. The first large-scale genotyping of more than 80,000 gene-based single nucleotide polymorphisms was performed in 2005 by Shimazaki et al. [29] in 920 Japanese T2DM patients. They identified an intronic variant, rs741301, of the engulfment and cell motility 1 (ELMO1) gene to be significantly associated with diabetic nephropathy. In vitro experiments suggested its role in the overaccumulation of extracellular matrix proteins and progression of glomerulosclerosis. Subsequently, a GWAS by Hanson et al. [30] used pooled DNA and validated the top signals using individual genotyping in 207 Pima Indian T2DM ESRD cases and controls. A variant in plasmacytoma variant translocation (PVT1) gene was suggestively associated with ESRD.
The first standard GWAS on diabetic nephropathy was published in 2009 by Pezzolesi et al. [31] and included the GoKinD study participants. They used two-stage GWAS with follow-up genotyping in 1,705 T1DM cases and controls. The four loci having a potential association signal of P<1.0×10-5 included FERM domain containing 3 (FRMD3), cysteinyl-tRNA synthetase (CARS), chimerin 2 (CHN2), and carboxypeptidase (CPVL). The rs1888747 variant in FRMD3 and rs451041 variant in CARS showed associations with time to onset of diabetic nephropathy in an independent cohort of DCCT/EDIC study with P<0.05. Craig et al. [32] reported a GWAS using pooled DNA of 932 T1DM GoKinD participants. They reported suggestive loci for ESRD on zinc finger, MIZ-type containing 1 (ZMIZ1) and musculin (MSC) genes as well as the association signals in six previously reported genetic loci of diabetic nephropathy (P≤0.0006). A relatively large-scale GWAS involving 3,393 African-American T2DM subjects was performed by McDonough et al. [33] in 2011. However, it was uncommon for African-Americans to have normal albuminuria after a diabetes duration of 10 years and the authors used nondiabetic controls for the control group. Therefore, analysis was performed to discriminate association signals between T2DM-associated ESRD, T2DM and all-cause ESRD using additional genotyping in African-Americans. Although none of the genetic variants reached genome-wide significance, several genes-including SAM and SH3 domain containing 1 (SASH1), ribosomal protein S12 gene (RPS12), and LIM kinase 2 (LIMK2)-were suggested as strong candidates for diabetic nephropathy in T2DM patients.
As previous studies were not fully powered for GWAS and the phenotype definition was different, a collaborative effort was made to conduct the Genetics of Nephropathy: An International Effort (GENIE) study on T1DM diabetic nephropathy [34]. This study was performed in two stages by Sandholm et al. [34]. In the first stage, a GWAS was meta-analyzed in three cohorts with a sample size of 5,783 subjects. In the second stage, de novo genotyping was performed in 5,873 participants from nine cohorts. The study was adequately powered and two variants were found associated with ESRD in AF4/FMR2 family, member 3 (AFF3) and in the intergenic region between repulsive guidance molecule family member A (RGMA) and multiple C2 domains, transmembrane 2 (MCTP2) that reached a genome wide significance threshold of P<5.0×10-8. In a GWAS on Finnish Diabetic Nephropathy study and GENIE consortium, Sandholm et al. [35] identified a gender-specific variant, rs4972593, which was associated with the risk of T1DM ESRD only in females. A GWAS was also conducted on urinary albumin excretion rate in 5,675 T1DM patients. This study identified rs2410601 in the intergenic region between pleckstrin and Sec7 domain containing 3 (PSD3) and SH2 domain containing 4A (SH2D4A) as the most significantly associated with albumin excretion rate (P=3.85×10-6) [36].
Among the nine GWASs, six included participants of European origin and one study each of Japanese subjects, Pima Indians and African-Americans. Only three studies were performed on T2DM, and the remaining six on T1DM subjects. Most of the studies included both ESRD and macroalbuminuria groups as cases. Only a few studies, including those from the GENIE consortium, had sufficient statistical power for GWAS and most of the earlier studies were limited in terms of sample size. Genetic variants in earlier reports, such as in ELMO1, were analyzed in subsequent studies and an association with diabetic nephropathy was confirmed in several, but not all, studies. Whether the pathophysiology of diabetic nephropathy differs between T1DM and T2DM patients remains unknown. Nonglycemic factors, such as insulin resistance and dyslipidemia, in T2DM may modulate the development of diabetic nephropathy and certain genetic risk factors could be involved in this process. Most of the well-powered GWASs were performed on T1DM patients and the genetic variants of diabetic nephropathy in T2DM patients should be elucidated.
Diabetic peripheral neuropathy is classified as generalized symmetric polyneuropathies and focal and multifocal neuropathies [38]. Diabetic sensorimotor polyneuropathy is one of the most common complications in diabetic patients with an estimated lifetime prevalence of up to 50% [38]. In 2009, the Toronto Consensus Panel on Diabetic Neuropathies updated its definition and diagnostic criteria for diabetic polyneuropathy [39]. Diagnosis of distal symmetric polyneuropathy is categorized into possible, probable, confirmed, and subclinical, according to the certainty based on symptoms and signs. The confirmation of neuropathy requires typical symptoms, signs, and positive nerve conduction studies.
Currently, only one GWAS on neuropathic pain in diabetic patients has been published (Table 3) [40]. Using the Genetics of Diabetes Audit and Research Tayside (GoDARTS) study, Meng et al. [40] performed a GWAS on 3,063 T2DM patients. The case control status was defined based on the prescription of medications frequently used for diabetic sensorimotor polyneuropathy, including duloxetine, gabapentin, pregabalin, capsaicin, and lidocaine patch. In a single-stage GWAS without follow-up genotyping, rs17428041 located in the intergenic region between the GDNF family receptor alpha 2 (GFRA2) and docking protein 2 (DOK2) was potentially associated with neuropathic pain (P=1.77×10-7). In addition, the narrow sense heritability of diabetic neuropathic pain was 11%, excluding the effect of gene-gene and gene-environment interactions. Further studies are required to replicate this finding and to identify additional genetic variants of diabetic neuropathy.
During the past several years, the identification of genetic risk factors for diabetic microvascular complications has improved. However, most of the studies were not fully powered for GWASs, with the exception of the GENIE study. Therefore, most of the results associated with the genetic risk factors were below the genome-wide significance threshold and inconsistent among studies. In addition, the definition of cases and controls differed, thereby introducing significant heterogeneity. Based on the findings reported, these genetic association results should be validated in other populations. In addition, a collaborative effort to harmonize phenotype definitions and to increase sample size is necessary.
Whether certain microvascular complications are caused by specific genetic risk factors, or common genetic risk factors are shared by different microvascular complications should be clarified. Additionally, a possible difference in genetic risk factors for microvascular complications between T1DM and T2DM patients should be explored. Whether confirmed genetic variants for T1DM or T2DM per se have significant effects on the development of microvascular complications remains unclear. Finally, a metabolic memory or legacy effect, as shown by the DCCT/EDIC trial, should be considered; this might be mediated by epigenetic change. Compared to T2DM, genetic studies on diabetic microvascular complications are still in the early stages and have further challenges to overcome. Further genetic studies of microvascular complications will enhance understanding of their pathogenesis and facilitate the development of effective preventive and therapeutic measures.
ACKNOWLEDGMENTS
This research was supported by a grant of Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea (grant number: HI14C0060).
References
1. Korean Diabetes Association. Diabetes Fact Sheet in Korea 2013 [Internet]. Seoul: Korean Diabetes Association;c2011. cited 2015 May 16. Available from: http://www.diabetes.or.kr/temp/diabetes_factsheet_2013111.pdf.
3. UK Prospective Diabetes Study (UKPDS) Group. Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33). Lancet. 1998; 352:837–853. PMID: 9742976.
4. ACCORD Study Group. ACCORD Eye Study Group. Chew EY, Ambrosius WT, Davis MD, Danis RP, Gangaputra S, Greven CM, Hubbard L, Esser BA, Lovato JF, Perdue LH, Goff DC Jr, Cushman WC, Ginsberg HN, Elam MB, Genuth S, Gerstein HC, Schubart U, Fine LJ. Effects of medical therapies on retinopathy progression in type 2 diabetes. N Engl J Med. 2010; 363:233–244. PMID: 20587587.
5. Ismail-Beigi F, Craven T, Banerji MA, Basile J, Calles J, Cohen RM, Cuddihy R, Cushman WC, Genuth S, Grimm RH Jr, Hamilton BP, Hoogwerf B, Karl D, Katz L, Krikorian A, O'Connor P, Pop-Busui R, Schubart U, Simmons D, Taylor H, Thomas A, Weiss D, Hramiak I. ACCORD trial group. Effect of intensive treatment of hyperglycaemia on microvascular outcomes in type 2 diabetes: an analysis of the ACCORD randomised trial. Lancet. 2010; 376:419–430. PMID: 20594588.

6. Looker HC, Nelson RG, Chew E, Klein R, Klein BE, Knowler WC, Hanson RL. Genome-wide linkage analyses to identify Loci for diabetic retinopathy. Diabetes. 2007; 56:1160–1166. PMID: 17395753.

7. Hietala K, Forsblom C, Summanen P, Groop PH. FinnDiane Study Group. Heritability of proliferative diabetic retinopathy. Diabetes. 2008; 57:2176–2180. PMID: 18443200.

8. Young BA, Maynard C, Boyko EJ. Racial differences in diabetic nephropathy, cardiovascular disease, and mortality in a national population of veterans. Diabetes Care. 2003; 26:2392–2399. PMID: 12882868.

9. Awata T, Inoue K, Kurihara S, Ohkubo T, Watanabe M, Inukai K, Inoue I, Katayama S. A common polymorphism in the 5'-untranslated region of the VEGF gene is associated with diabetic retinopathy in type 2 diabetes. Diabetes. 2002; 51:1635–1639. PMID: 11978667.

10. Abhary S, Burdon KP, Laurie KJ, Thorpe S, Landers J, Goold L, Lake S, Petrovsky N, Craig JE. Aldose reductase gene polymorphisms and diabetic retinopathy susceptibility. Diabetes Care. 2010; 33:1834–1836. PMID: 20424224.

11. Tong Z, Yang Z, Patel S, Chen H, Gibbs D, Yang X, Hau VS, Kaminoh Y, Harmon J, Pearson E, Buehler J, Chen Y, Yu B, Tinkham NH, Zabriskie NA, Zeng J, Luo L, Sun JK, Prakash M, Hamam RN, Tonna S, Constantine R, Ronquillo CC, Sadda S, Avery RL, Brand JM, London N, Anduze AL, King GL, Bernstein PS, Watkins S. Genetics of Diabetes and Diabetic Complication Study Group. Jorde LB, Li DY, Aiello LP, Pollak MR, Zhang K. Promoter polymorphism of the erythropoietin gene in severe diabetic eye and kidney complications. Proc Natl Acad Sci U S A. 2008; 105:6998–7003. PMID: 18458324.

12. Wang F, Fang Q, Yu N, Zhao D, Zhang Y, Wang J, Wang Q, Zhou X, Cao X, Fan X. Association between genetic polymorphism of the angiotensin-converting enzyme and diabetic nephropathy: a meta-analysis comprising 26,580 subjects. J Renin Angiotensin Aldosterone Syst. 2012; 13:161–174. PMID: 21810896.

13. Ma RC, Tam CH, Wang Y, Luk AO, Hu C, Yang X, Lam V, Chan AW, Ho JS, Chow CC, Tong PC, Jia W, Ng MC, So WY, Chan JC. Genetic variants of the protein kinase C-beta 1 gene and development of end-stage renal disease in patients with type 2 diabetes. JAMA. 2010; 304:881–889. PMID: 20736472.
14. Sobrin L, Green T, Sim X, Jensen RA, Tai ES, Tay WT, Wang JJ, Mitchell P, Sandholm N, Liu Y, Hietala K, Iyengar SK. Family Investigation of Nephropathy and Diabetes-Eye Research Group. Brooks M, Buraczynska M, Van Zuydam N, Smith AV, Gudnason V, Doney AS, Morris AD, Leese GP, Palmer CN. Wellcome Trust Case Control Consortium 2. Swaroop A, Taylor HA Jr, Wilson JG, Penman A, Chen CJ, Groop PH, Saw SM, Aung T, Klein BE, Rotter JI, Siscovick DS, Cotch MF, Klein R, Daly MJ, Wong TY. Candidate gene association study for diabetic retinopathy in persons with type 2 diabetes: the Candidate gene Association Resource (CARe). Invest Ophthalmol Vis Sci. 2011; 52:7593–7602. PMID: 21873659.

15. International HapMap Consortium. Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Shen Y, Sun W, Wang H, Wang Y, Wang Y, Xiong X, Xu L, Waye MM, Tsui SK, Xue H, Wong JT, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallee C, Verner A, Hudson TJ, Kwok PY, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui LC, Mak W, Song YQ, Tam PK, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, Deloukas P, Bird CP, Delgado M, Dermitzakis ET, Gwilliam R, Hunt S, Morrison J, Powell D, Stranger BE, Whittaker P, Bentley DR, Daly MJ, de Bakker PI, Barrett J, Chretien YR, Maller J, McCarroll S, Patterson N, Pe'er I, Price A, Purcell S, Richter DJ, Sabeti P, Saxena R, Schaffner SF, Sham PC, Varilly P, Altshuler D, Stein LD, Krishnan L, Smith AV, Tello-Ruiz MK, Thorisson GA, Chakravarti A, Chen PE, Cutler DJ, Kashuk CS, Lin S, Abecasis GR, Guan W, Li Y, Munro HM, Qin ZS, Thomas DJ, McVean G, Auton A, Bottolo L, Cardin N, Eyheramendy S, Freeman C, Marchini J, Myers S, Spencer C, Stephens M, Donnelly P, Cardon LR, Clarke G, Evans DM, Morris AP, Weir BS, Tsunoda T, Mullikin JC, Sherry ST, Feolo M, Skol A, Zhang H, Zeng C, Zhao H, Matsuda I, Fukushima Y, Macer DR, Suda E, Rotimi CN, Adebamowo CA, Ajayi I, Aniagwu T, Marshall PA, Nkwodimmah C, Royal CD, Leppert MF, Dixon M, Peiffer A, Qiu R, Kent A, Kato K, Niikawa N, Adewole IF, Knoppers BM, Foster MW, Clayton EW, Watkin J, Gibbs RA, Belmont JW, Muzny D, Nazareth L, Sodergren E, Weinstock GM, Wheeler DA, Yakub I, Gabriel SB, Onofrio RC, Richter DJ, Ziaugra L, Birren BW, Daly MJ, Altshuler D, Wilson RK, Fulton LL, Rogers J, Burton J, Carter NP, Clee CM, Griffiths M, Jones MC, McLay K, Plumb RW, Ross MT, Sims SK, Willey DL, Chen Z, Han H, Kang L, Godbout M, Wallenburg JC, L'Archeveque P, Bellemare G, Saeki K, Wang H, An D, Fu H, Li Q, Wang Z, Wang R, Holden AL, Brooks LD, McEwen JE, Guyer MS, Wang VO, Peterson JL, Shi M, Spiegel J, Sung LM, Zacharia LF, Collins FS, Kennedy K, Jamieson R, Stewart J. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007; 449:851–861. PMID: 17943122.
16. Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009; 461:747–753. PMID: 19812666.

17. Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007; 445:881–885. PMID: 17293876.

18. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium. Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium. South Asian Type 2 Diabetes (SAT2D) Consortium. Mexican American Type 2 Diabetes (MAT2D) Consortium. Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium. Mahajan A, Go MJ, Zhang W, Below JE, Gaulton KJ, Ferreira T, Horikoshi M, Johnson AD, Ng MC, Prokopenko I, Saleheen D, Wang X, Zeggini E, Abecasis GR, Adair LS, Almgren P, Atalay M, Aung T, Baldassarre D, Balkau B, Bao Y, Barnett AH, Barroso I, Basit A, Been LF, Beilby J, Bell GI, Benediktsson R, Bergman RN, Boehm BO, Boerwinkle E, Bonnycastle LL, Burtt N, Cai Q, Campbell H, Carey J, Cauchi S, Caulfield M, Chan JC, Chang LC, Chang TJ, Chang YC, Charpentier G, Chen CH, Chen H, Chen YT, Chia KS, Chidambaram M, Chines PS, Cho NH, Cho YM, Chuang LM, Collins FS, Cornelis MC, Couper DJ, Crenshaw AT, van Dam RM, Danesh J, Das D, de Faire U, Dedoussis G, Deloukas P, Dimas AS, Dina C, Doney AS, Donnelly PJ, Dorkhan M, van Duijn C, Dupuis J, Edkins S, Elliott P, Emilsson V, Erbel R, Eriksson JG, Escobedo J, Esko T, Eury E, Florez JC, Fontanillas P, Forouhi NG, Forsen T, Fox C, Fraser RM, Frayling TM, Froguel P, Frossard P, Gao Y, Gertow K, Gieger C, Gigante B, Grallert H, Grant GB, Grrop LC, Groves CJ, Grundberg E, Guiducci C, Hamsten A, Han BG, Hara K, Hassanali N, Hattersley AT, Hayward C, Hedman AK, Herder C, Hofman A, Holmen OL, Hovingh K, Hreidarsson AB, Hu C, Hu FB, Hui J, Humphries SE, Hunt SE, Hunter DJ, Hveem K, Hydrie ZI, Ikegami H, Illig T, Ingelsson E, Islam M, Isomaa B, Jackson AU, Jafar T, James A, Jia W, Jockel KH, Jonsson A, Jowett JB, Kadowaki T, Kang HM, Kanoni S, Kao WH, Kathiresan S, Kato N, Katulanda P, Keinanen-Kiukaanniemi KM, Kelly AM, Khan H, Khaw KT, Khor CC, Kim HL, Kim S, Kim YJ, Kinnunen L, Klopp N, Kong A, Korpi-Hyovalti E, Kowlessur S, Kraft P, Kravic J, Kristensen MM, Krithika S, Kumar A, Kumate J, Kuusisto J, Kwak SH, Laakso M, Lagou V, Lakka TA, Langenberg C, Langford C, Lawrence R, Leander K, Lee JM, Lee NR, Li M, Li X, Li Y, Liang J, Liju S, Lim WY, Lind L, Lindgren CM, Lindholm E, Liu CT, Liu JJ, Lobbens S, Long J, Loos RJ, Lu W, Luan J, Lyssenko V, Ma RC, Maeda S, Magi R, Mannisto S, Matthews DR, Meigs JB, Melander O, Metspalu A, Meyer J, Mirza G, Mihailov E, Moebus S, Mohan V, Mohlke KL, Morris AD, Muhleisen TW, Muller-Nurasyid M, Musk B, Nakamura J, Nakashima E, Navarro P, Ng PK, Nica AC, Nilsson PM, Njolstad I, Nothen MM, Ohnaka K, Ong TH, Owen KR, Palmer CN, Pankow JS, Park KS, Parkin M, Pechlivanis S, Pedersen NL, Peltonen L, Perry JR, Peters A, Pinidiyapathirage JM, Platou CG, Potter S, Price JF, Qi L, Radha V, Rallidis L, Rasheed A, Rathman W, Rauramaa R, Raychaudhuri S, Rayner NW, Rees SD, Rehnberg E, Ripatti S, Robertson N, Roden M, Rossin EJ, Rudan I, Rybin D, Saaristo TE, Salomaa V, Saltevo J, Samuel M, Sanghera DK, Saramies J, Scott J, Scott LJ, Scott RA, Segre AV, Sehmi J, Sennblad B, Shah N, Shah S, Shera AS, Shu XO, Shuldiner AR, Sigurdsson G, Sijbrands E, Silveira A, Sim X, Sivapalaratnam S, Small KS, So WY, Stancakova A, Stefansson K, Steinbach G, Steinthorsdottir V, Stirrups K, Strawbridge RJ, Stringham HM, Sun Q, Suo C, Syvanen AC, Takayanagi R, Takeuchi F, Tay WT, Teslovich TM, Thorand B, Thorleifsson G, Thorsteinsdottir U, Tikkanen E, Trakalo J, Tremoli E, Trip MD, Tsai FJ, Tuomi T, Tuomilehto J, Uitterlinden AG, Valladares-Salgado A, Vedantam S, Veglia F, Voight BF, Wang C, Wareham NJ, Wennauer R, Wickremasinghe AR, Wilsgaard T, Wilson JF, Wiltshire S, Winckler W, Wong TY, Wood AR, Wu JY, Wu Y, Yamamoto K, Yamauchi T, Yang M, Yengo L, Yokota M, Young R, Zabaneh D, Zhang F, Zhang R, Zheng W, Zimmet PZ, Altshuler D, Bowden DW, Cho YS, Cox NJ, Cruz M, Hanis CL, Kooner J, Lee JY, Seielstad M, Teo YY, Boehnke M, Parra EJ, Chambers JC, Tai ES, McCarthy MI, Morris AP. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014; 46:234–244. PMID: 24509480.

19. Hivert MF, Vassy JL, Meigs JB. Susceptibility to type 2 diabetes mellitus: from genes to prevention. Nat Rev Endocrinol. 2014; 10:198–205. PMID: 24535206.
20. Early Treatment Diabetic Retinopathy Study Research Group. Grading diabetic retinopathy from stereoscopic color fundus photographs: an extension of the modified Airlie House classification. ETDRS report number 10. Ophthalmology. 1991; 98(5 Suppl):786–806. PMID: 2062513.
21. Klein R, Klein BE, Moss SE, Davis MD, DeMets DL. The Wisconsin epidemiologic study of diabetic retinopathy. II. Prevalence and risk of diabetic retinopathy when age at diagnosis is less than 30 years. Arch Ophthalmol. 1984; 102:520–526. PMID: 6367724.
22. Fu YP, Hallman DM, Gonzalez VH, Klein BE, Klein R, Hayes MG, Cox NJ, Bell GI, Hanis CL. Identification of diabetic retinopathy genes through a genome-wide association study among Mexican-Americans from Starr County, Texas. J Ophthalmol. 2010; 2010:pii: 861291.

23. Huang YC, Lin JM, Lin HJ, Chen CC, Chen SY, Tsai CH, Tsai FJ. Genome-wide association study of diabetic retinopathy in a Taiwanese population. Ophthalmology. 2011; 118:642–648. PMID: 21310492.

24. Grassi MA, Tikhomirov A, Ramalingam S, Below JE, Cox NJ, Nicolae DL. Genome-wide meta-analysis for severe diabetic retinopathy. Hum Mol Genet. 2011; 20:2472–2481. PMID: 21441570.

25. Sheu WH, Kuo JZ, Lee IT, Hung YJ, Lee WJ, Tsai HY, Wang JS, Goodarzi MO, Klein R, Klein BE, Ipp E, Lin SY, Guo X, Hsieh CH, Taylor KD, Fu CP, Rotter JI, Chen YD. Genome-wide association study in a Chinese population with diabetic retinopathy. Hum Mol Genet. 2013; 22:3165–3173. PMID: 23562823.

26. Awata T, Yamashita H, Kurihara S, Morita-Ohkubo T, Miyashita Y, Katayama S, Mori K, Yoneya S, Kohda M, Okazaki Y, Maruyama T, Shimada A, Yasuda K, Nishida N, Tokunaga K, Koike A. Correction: a genome-wide association study for diabetic retinopathy in a Japanese population: potential association with a long intergenic non-coding RNA. PLoS One. 2015; 10:e0126789. PMID: 25910184.

27. Perkovic V, Heerspink HL, Chalmers J, Woodward M, Jun M, Li Q, MacMahon S, Cooper ME, Hamet P, Marre M, Mogensen CE, Poulter N, Mancia G, Cass A, Patel A, Zoungas S. ADVANCE Collaborative Group. Intensive glucose control improves kidney outcomes in patients with type 2 diabetes. Kidney Int. 2013; 83:517–523. PMID: 23302714.

28. The Diabetes Control and Complications Trial Research Group. The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. N Engl J Med. 1993; 329:977–986. PMID: 8366922.
29. Shimazaki A, Kawamura Y, Kanazawa A, Sekine A, Saito S, Tsunoda T, Koya D, Babazono T, Tanaka Y, Matsuda M, Kawai K, Iiizumi T, Imanishi M, Shinosaki T, Yanagimoto T, Ikeda M, Omachi S, Kashiwagi A, Kaku K, Iwamoto Y, Kawamori R, Kikkawa R, Nakajima M, Nakamura Y, Maeda S. Genetic variations in the gene encoding ELMO1 are associated with susceptibility to diabetic nephropathy. Diabetes. 2005; 54:1171–1178. PMID: 15793258.

30. Hanson RL, Craig DW, Millis MP, Yeatts KA, Kobes S, Pearson JV, Lee AM, Knowler WC, Nelson RG, Wolford JK. Identification of PVT1 as a candidate gene for end-stage renal disease in type 2 diabetes using a pooling-based genome-wide single nucleotide polymorphism association study. Diabetes. 2007; 56:975–983. PMID: 17395743.

31. Pezzolesi MG, Poznik GD, Mychaleckyj JC, Paterson AD, Barati MT, Klein JB, Ng DP, Placha G, Canani LH, Bochenski J, Waggott D, Merchant ML, Krolewski B, Mirea L, Wanic K, Katavetin P, Kure M, Wolkow P, Dunn JS, Smiles A, Walker WH, Boright AP, Bull SB. DCCT/EDIC Research Group. Doria A, Rogus JJ, Rich SS, Warram JH, Krolewski AS. Genome-wide association scan for diabetic nephropathy susceptibility genes in type 1 diabetes. Diabetes. 2009; 58:1403–1410. PMID: 19252134.

32. Craig DW, Millis MP, DiStefano JK. Genome-wide SNP genotyping study using pooled DNA to identify candidate markers mediating susceptibility to end-stage renal disease attributed to type 1 diabetes. Diabet Med. 2009; 26:1090–1098. PMID: 19929986.

33. McDonough CW, Palmer ND, Hicks PJ, Roh BH, An SS, Cooke JN, Hester JM, Wing MR, Bostrom MA, Rudock ME, Lewis JP, Talbert ME, Blevins RA, Lu L, Ng MC, Sale MM, Divers J, Langefeld CD, Freedman BI, Bowden DW. A genome-wide association study for diabetic nephropathy genes in African Americans. Kidney Int. 2011; 79:563–572. PMID: 21150874.

34. Sandholm N, Salem RM, McKnight AJ, Brennan EP, Forsblom C, Isakova T, McKay GJ, Williams WW, Sadlier DM, Makinen VP, Swan EJ, Palmer C, Boright AP, Ahlqvist E, Deshmukh HA, Keller BJ, Huang H, Ahola AJ, Fagerholm E, Gordin D, Harjutsalo V, He B, Heikkila O, Hietala K, Kyto J, Lahermo P, Lehto M, Lithovius R, Osterholm AM, Parkkonen M, Pitkaniemi J, Rosengard-Barlund M, Saraheimo M, Sarti C, Soderlund J, Soro-Paavonen A, Syreeni A, Thorn LM, Tikkanen H, Tolonen N, Tryggvason K, Tuomilehto J, Waden J, Gill GV, Prior S, Guiducci C, Mirel DB, Taylor A, Hosseini SM. DCCT/EDIC Research Group. Parving HH, Rossing P, Tarnow L, Ladenvall C, Alhenc-Gelas F, Lefebvre P, Rigalleau V, Roussel R, Tregouet DA, Maestroni A, Maestroni S, Falhammar H, Gu T, Mollsten A, Cimponeriu D, Ioana M, Mota M, Mota E, Serafinceanu C, Stavarachi M, Hanson RL, Nelson RG, Kretzler M, Colhoun HM, Panduru NM, Gu HF, Brismar K, Zerbini G, Hadjadj S, Marre M, Groop L, Lajer M, Bull SB, Waggott D, Paterson AD, Savage DA, Bain SC, Martin F, Hirschhorn JN, Godson C, Florez JC, Groop PH, Maxwell AP. New susceptibility loci associated with kidney disease in type 1 diabetes. PLoS Genet. 2012; 8:e1002921. PMID: 23028342.

35. Sandholm N, McKnight AJ, Salem RM, Brennan EP, Forsblom C, Harjutsalo V, Makinen VP, McKay GJ, Sadlier DM, Williams WW, Martin F, Panduru NM, Tarnow L, Tuomilehto J, Tryggvason K, Zerbini G, Comeau ME, Langefeld CD, Consortium F, Godson C, Hirschhorn JN, Maxwell AP, Florez JC, Groop PH. FinnDiane Study Group and the GENIE Consortium. Chromosome 2q31.1 associates with ESRD in women with type 1 diabetes. J Am Soc Nephrol. 2013; 24:1537–1543. PMID: 24029427.

36. Sandholm N, Forsblom C, Makinen VP, McKnight AJ, Osterholm AM, He B, Harjutsalo V, Lithovius R, Gordin D, Parkkonen M, Saraheimo M, Thorn LM, Tolonen N, Waden J, Tuomilehto J, Lajer M, Ahlqvist E, Mollsten A, Marcovecchio ML, Cooper J, Dunger D, Paterson AD, Zerbini G, Groop L, Consortium S, Tarnow L, Maxwell AP, Tryggvason K, Groop PH. SUMMIT Consortium, FinnDiane Study Group. Genome-wide association study of urinary albumin excretion rate in patients with type 1 diabetes. Diabetologia. 2014; 57:1143–1153. PMID: 24595857.

37. Germain M, Pezzolesi MG, Sandholm N, McKnight AJ, Susztak K, Lajer M, Forsblom C, Marre M, Parving HH, Rossing P, Toppila I, Skupien J, Roussel R, Ko YA, Ledo N, Folkersen L, Civelek M, Maxwell AP, Tregouet DA, Groop PH, Tarnow L, Hadjadj S. SORBS1 gene, a new candidate for diabetic nephropathy: results from a multi-stage genome-wide association study in patients with type 1 diabetes. Diabetologia. 2015; 58:543–548. PMID: 25476525.

38. Tesfaye S, Boulton AJ, Dyck PJ, Freeman R, Horowitz M, Kempler P, Lauria G, Malik RA, Spallone V, Vinik A, Bernardi L, Valensi P. Toronto Diabetic Neuropathy Expert Group. Diabetic neuropathies: update on definitions, diagnostic criteria, estimation of severity, and treatments. Diabetes Care. 2010; 33:2285–2293. PMID: 20876709.

39. Dyck PJ, Albers JW, Andersen H, Arezzo JC, Biessels GJ, Bril V, Feldman EL, Litchy WJ, O'Brien PC, Russell JW. Toronto Expert Panel on Diabetic Neuropathy. Diabetic polyneuropathies: update on research definition, diagnostic criteria and estimation of severity. Diabetes Metab Res Rev. 2011; 27:620–628. PMID: 21695763.

40. Meng W, Deshmukh HA, van Zuydam NR, Liu Y, Donnelly LA, Zhou K. Wellcome Trust Case Control Consortium 2 (WTCCC2). Surrogate Markers for Micro- and Macro-Vascular Hard Endpoints for Innovative Diabetes Tools (SUMMIT) Study Group. Morris AD, Colhoun HM, Palmer CN, Smith BH. A genome-wide association study suggests an association of Chr8p213 (GFRA2) with diabetic neuropathic pain. Eur J Pain. 2015; 19:392–399. PMID: 24974787.
Table 1
Genome-Wide Association Study on Diabetic Retinopathy
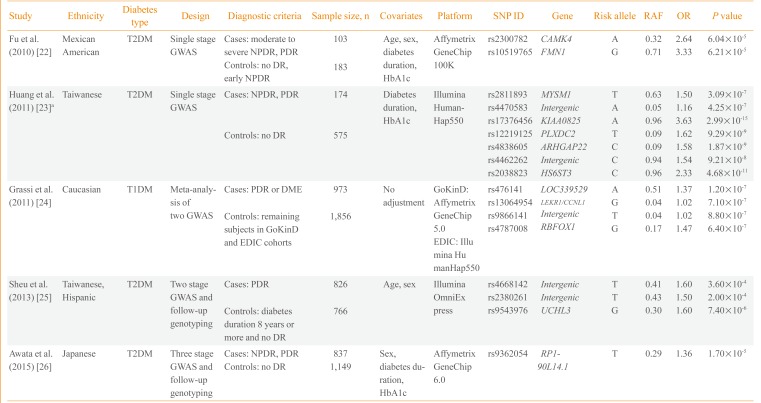
| Study | Ethnicity | Diabetes type | Design | Diagnostic criteria | Sample size, n | Covariates | Platform | SNP ID | Gene | Risk allele | RAF | OR | P value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fu et al. (2010) [22] | Mexican | T2DM | Single stage GWAS | Cases: moderate to severe NPDR, PDR | 103 | Age, sex, diabetes duration, HbA1c | Affymetrix GeneChip 100K | rs2300782 | CAMK4 | A | 0.32 | 2.64 | 6.04×10-5 |
| American | rs10519765 | FMN1 | G | 0.71 | 3.33 | 6.21×10-5 | |||||||
| Controls: no DR, early NPDR | 183 | ||||||||||||
| Huang et al. (2011) [23]a | Taiwanese | T2DM | Single stage GWAS | Cases: NPDR, PDR | 174 | Diabetes duration, HbA1c | Illumina HumanHap550 | rs2811893 | MYSM1 | T | 0.63 | 1.50 | 3.09×10-7 |
| rs4470583 | Intergenic | A | 0.05 | 1.16 | 4.25×10-7 | ||||||||
| rs17376456 | KIAA0825 | A | 0.96 | 3.63 | 2.99×10-15 | ||||||||
| Controls: no DR | 575 | rs12219125 | PLXDC2 | T | 0.09 | 1.62 | 9.29×10-9 | ||||||
| rs4838605 | ARHGAP22 | C | 0.09 | 1.58 | 1.87×10-9 | ||||||||
| rs4462262 | Intergenic | C | 0.94 | 1.54 | 9.21×10-8 | ||||||||
| rs2038823 | HS6ST3 | C | 0.96 | 2.33 | 4.68×10-11 | ||||||||
| Grassi et al. (2011) [24] | Caucasian | T1DM | Meta-analysis of two GWAS | Cases: PDR or DME | 973 | No adjustment | GoKinD: Affymetrix GeneChip 5.0 | rs476141 | LOC339529 | A | 0.51 | 1.37 | 1.20×10-7 |
| rs13064954 | LEKR1/CCNL1 | G | 0.04 | 1.02 | 7.10×10-7 | ||||||||
| Controls: remaining subjects in GoKinD and EDIC cohorts | 1,856 | rs9866141 | Intergenic | T | 0.04 | 1.02 | 8.80×10-7 | ||||||
| rs4787008 | RBFOX1 | G | 0.17 | 1.47 | 6.40×10-7 | ||||||||
| EDIC: Illumina HumanHap550 | |||||||||||||
| Sheu et al. (2013) [25] | Taiwanese, Hispanic | T2DM | Two stage GWAS and follow-up genotyping | Cases: PDR | 826 | Age, sex | Illumina OmniExpress | rs4668142 | Intergenic | T | 0.41 | 1.60 | 3.60×10-4 |
| rs2380261 | Intergenic | T | 0.43 | 1.50 | 2.00×10-4 | ||||||||
| Controls: diabetes duration 8 years or more and no DR | 766 | rs9543976 | UCHL3 | G | 0.30 | 1.60 | 7.40×10-6 | ||||||
| Awata et al. (2015) [26] | Japanese | T2DM | Three stage GWAS and follow-up genotyping | Cases: NPDR, PDR | 837 | Sex, diabetes duration, HbA1c | Affymetrix GeneChip 6.0 | rs9362054 | RP1-90L14.1 | T | 0.29 | 1.36 | 1.70×10-5 |
| Controls: no DR | 1,149 |
SNP, single nucleotide polymorphism; RAF, risk allele frequency in controls; OR, odds ratio; T2DM, type 2 diabetes mellitus; GWAS, genome-wide association study; NPDR, nonproliferative diabetic retinopathy; PDR, proliferative diabetic retinopathy; DR, diabetic retinopathy; HbA1c, hemoglobin A1c; T1DM, type 1 diabetes mellitus; DME, diabetic macular edema; GoKinD, Genetics of Kidneys in Diabetes; EDIC, Epidemiology of Diabetes Interventions and Complications.
aOR are from dominant model and P values are from the lowest among six genetics models.
Table 2
Genome-Wide Association Study on Diabetic Nephropathy
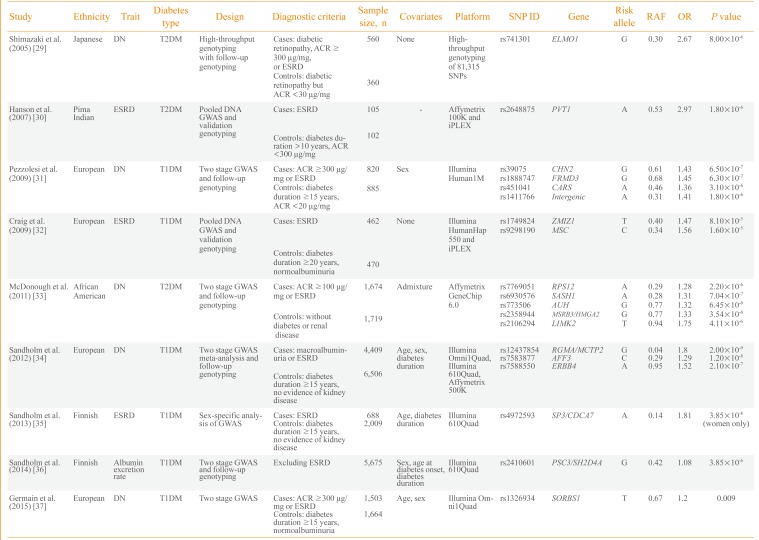
| Study | Ethnicity | Trait | Diabetes type | Design | Diagnostic criteria | Sample size, n | Covariates | Platform | SNP ID | Gene | Risk allele | RAF | OR | P value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Shimazaki et al. (2005) [29] | Japanese | DN | T2DM | High-throughput genotyping with follow-up genotyping | Cases: diabetic retinopathy, ACR ≥300 µg/mg, or ESRD | 560 | None | High-throughput genotyping of 81,315 SNPs | rs741301 | ELMO1 | G | 0.30 | 2.67 | 8.00×10-6 |
| Controls: diabetic retinopathy but ACR <30 µg/mg | 360 | |||||||||||||
| Hanson et al. (2007) [30] | Pima | ESRD | T2DM | Pooled DNA GWAS and validation genotyping | Cases: ESRD | 105 | - | Affymetrix 100K and iPLEX | rs2648875 | PVT1 | A | 0.53 | 2.97 | 1.80×10-6 |
| Indian | ||||||||||||||
| Controls: diabetes duration >10 years, ACR <300 µg/mg | 102 | |||||||||||||
| Pezzolesi et al. (2009) [31] | European | DN | T1DM | Two stage GWAS and follow-up genotyping | Cases: ACR ≥300 µg/mg or ESRD | 820 | Sex | Illumina Human1M | rs39075 | CHN2 | G | 0.61 | 1.43 | 6.50×10-7 |
| rs1888747 | FRMD3 | G | 0.68 | 1.45 | 6.30×10-7 | |||||||||
| Controls: diabetes duration ≥15 years, ACR <20 µg/mg | 885 | rs451041 | CARS | A | 0.46 | 1.36 | 3.10×10-6 | |||||||
| rs1411766 | Intergenic | A | 0.31 | 1.41 | 1.80×10-6 | |||||||||
| Craig et al. (2009) [32] | European | ESRD | T1DM | Pooled DNA GWAS and validation genotyping | Cases: ESRD | 462 | None | Illumina HumanHap 550 and iPLEX | rs1749824 | ZMIZ1 | T | 0.40 | 1.47 | 8.10×10-5 |
| rs9298190 | MSC | C | 0.34 | 1.56 | 1.60×10-5 | |||||||||
| Controls: diabetes duration ≥20 years, normoalbuminuria | 470 | |||||||||||||
| McDonough et al. (2011) [33] | African | DN | T2DM | Two stage GWAS and follow-up genotyping | Cases: ACR ≥100 µg/mg or ESRD | 1,674 | Admixture | Affymetrix GeneChip 6.0 | rs7769051 | RPS12 | A | 0.29 | 1.28 | 2.20×10-6 |
| American | rs6930576 | SASH1 | A | 0.28 | 1.31 | 7.04×10-7 | ||||||||
| rs773506 | AUH | G | 0.77 | 1.32 | 6.45×10-6 | |||||||||
| Controls: without diabetes or renal disease | 1,719 | rs2358944 | MSRB3/HMGA2 | G | 0.77 | 1.33 | 3.54×10-6 | |||||||
| rs2106294 | LIMK2 | T | 0.94 | 1.75 | 4.11×10-6 | |||||||||
| Sandholm et al. (2012) [34] | European | DN | T1DM | Two stage GWAS meta-analysis and follow-up genotyping | Cases: macroalbuminuria or ESRD | 4,409 | Age, sex, diabetes duration | Illumina Omni1Quad, Illumina 610Quad, Affymetrix 500K | rs12437854 | RGMA/MCTP2 | G | 0.04 | 1.8 | 2.00×10-9 |
| rs7583877 | AFF3 | C | 0.29 | 1.29 | 1.20×10-8 | |||||||||
| rs7588550 | ERBB4 | A | 0.95 | 1.52 | 2.10×10-7 | |||||||||
| Controls: diabetes duration ≥15 years, no evidence of kidney disease | 6,506 | |||||||||||||
| Sandholm et al. (2013) [35] | Finnish | ESRD | T1DM | Sex-specific analysis of GWAS | Cases: ESRD | 688 | Age, diabetes duration | Illumina 610Quad | rs4972593 | SP3/CDCA7 | A | 0.14 | 1.81 | 3.85×10-8 (women only) |
| Controls: diabetes duration ≥15 years, no evidence of kidney disease | 2,009 | |||||||||||||
| Sandholm et al. (2014) [36] | Finnish | Albumin excretion rate | T1DM | Two stage GWAS and follow-up genotyping | Excluding ESRD | 5,675 | Sex, age at diabetes onset, diabetes duration | Illumina 610Quad | rs2410601 | PSC3/SH2D4A | G | 0.42 | 1.08 | 3.85×10-6 |
| Germain et al. (2015) [37] | European | DN | T1DM | Two stage GWAS | Cases: ACR ≥300 µg/mg or ESRD | 1,503 | Age, sex | Illumina Omni1Quad | rs1326934 | SORBS1 | T | 0.67 | 1.2 | 0.009 |
| Controls: diabetes duration ≥15 years, normoalbuminuria | 1,664 |
Table 3
Genome-Wide Association Study on Diabetic Neuropathy
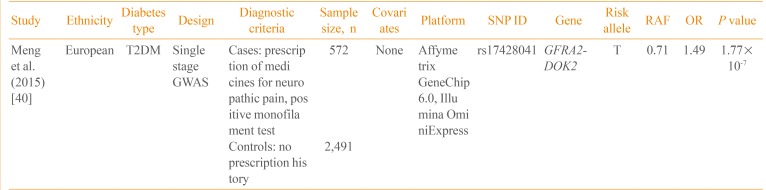
| Study | Ethnicity | Diabetes type | Design | Diagnostic criteria | Sample size, n | Covariates | Platform | SNP ID | Gene | Risk allele | RAF | OR | P value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Meng et al. (2015) [40] | European | T2DM | Single stage GWAS | Cases: prescription of medicines for neuropathic pain, positive monofilament test | 572 | None | Affymetrix GeneChip 6.0, Illumina OminiExpress | rs17428041 | GFRA2-DOK2 | T | 0.71 | 1.49 | 1.77×10-7 |
| Controls: no prescription history | 2,491 |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download