Abstract
Background
In mammals, the CLOCK/BMAL1 heterodimer is a key transcription factor complex that drives the cyclic expression of clock-controlled genes involved in various physiological functions and behavioral consequences. Recently, a growing number of studies have reported a molecular link between the circadian clock and metabolism. In the present study, we explored the regulatory effects of SIRTUIN1 (SIRT1), an NAD+-dependent deacetylase, on CLOCK/BMAL1-mediated clock gene expression.
Methods
To investigate the interaction between SIRT1 and CLOCK/BMAL1, we conducted bimolecular fluorescence complementation (BiFC) analyses supplemented with immunocytochemistry assays. BiFC experiments employing deletion-specific mutants of BMAL1 were used to elucidate the specific domains that are necessary for the SIRT1-BMAL1 interaction. Additionally, luciferase reporter assays were used to delineate the effects of SIRT1 on circadian gene expression.
Results
BiFC analysis revealed that SIRT1 interacted with both CLOCK and BMAL1 in most cell nuclei. As revealed by BiFC assays using various BMAL1 deletion mutants, the PAS-B domain of BMAL1 was essential for interaction with SIRT1. Activation of SIRT1 with resveratrol did not exert any significant change on the interaction with the CLOCK/BMAL1 complex. However, promoter analysis using Per1-Luc and Ebox-Luc reporters showed that SIRT1 significantly downregulated both promoter activities. This inhibitory effect was intensified by treatment with resveratrol, indicating a role for SIRT1 and its activator in CLOCK/BMAL1-mediated transcription of clock genes.
Circadian rhythm is an approximately 24-hour biological cycle that allows organisms to adapt their physiology and behavior to the day/night cycle. In mammals, the circadian timing system is controlled by a central pacemaker in the suprachiasmatic nucleus (SCN) of the hypothalamus and subsidiary oscillators in most peripheral tissues [1]. The light/dark cycle is the predominant zeitgeber (timing cue) for the SCN, whereas cyclic feeding behavior is a strong zeitgeber for many peripheral tissue clocks [2]. The central clock harmonizes peripheral oscillators via various kinds of outputs, including neural and humoral signals, rest-activity rhythms, and body temperature. Thus, some outputs of the SCN work as direct inputs to the peripheral tissues, such as the feeding-fasting cycle [2].
The autonomous and self-sustainable nature of circadian timing is largely dependent on the molecular circadian clockwork. The molecular mechanism underlying the mammalian circadian clock consists of a transcription-translation feedback loop involving CLOCK and BMAL1, which recognize E-box elements. The CLOCK/BMAL1 heterodimer activates the transcription of period (Per1, 2, and 3) and cryptochrome (Cry1 and 2), leading to the subsequent repression of CLOCK/BMAL1 activity by CRY and PER proteins [3]. An additional feedback loop involves the transcriptional regulation of Bmal1 by RAR-related orphan receptor α (RORα) and Rev-erb α [4,5].
Recently, accumulating evidence suggests a strong interplay between the circadian clock and metabolism [6,7,8]. Indeed, the cellular DNA-binding activity of CLOCK/BMAL1 is strongly influenced by the ratio of reduced to oxidized nicotinamide adenine dinucleotide (NAD) cofactors, indicating that the cellular metabolic state regulates the molecular clock. Furthermore, SIRTUIN1 (SIRT1), an NAD+-dependent deacetylase, is known to regulate the circadian clock circuitry [9,10,11,12]. SIRT1, the closest mammalian homologue of yeast Sir2, regulates a variety of cellular processes, including cell survival, development, inflammation, and metabolism [13,14]. The SIRT1 catalytic reaction involves the breakdown of one NAD+ molecule for the deacetylation of acetyl lysine and the generation of nicotinamide and O-acetyl-ADP-ribose. SIRT1 is known to deacetylate not only histones, but also several transcriptional regulatory proteins that control metabolism [15,16,17]. Recent reports have shown that SIRT1 is a component of CLOCK/BMAL1 transcription complexes and affects the expression of clock genes [9,10].
Recently, there has been interest in the identification of SIRT1 activators and activating compounds. For instance, resveratrol, a natural small polyphenol found in red grapes and wine, is well-known as a SIRT1 activator [18]. Accordingly, resveratrol is a subject of great interest since it was shown to exert beneficial effects on glucose and lipid metabolism. Moreover, resveratrol was shown to extend life span in rodents [19,20]. Despite the close involvement of SIRT1 in the circadian clock and metabolism, the precise mechanism of SIRT1 activation by resveratrol remains unclear [21]. In the present study, we attempt to visualize the interaction of SIRT1 with CLOCK/BMAL1 in a native cellular context using a bimolecular fluorescence complementation (BiFC) analysis.
Human SIRT1 was amplified from HA-FLAG-tagged human SIRT1 (a kind gift from Gad Asher, University of Geneva, Switzerland) by polymerase chain reaction (PCR) using SIRT1-specific primers (forward, 5'-GATATCATGGCGGACGAGGCGGCCC; reverse, 3'-GTCGACTGATTTGTTTGATGGATAGTTC). It was then subcloned into a cDNA encoding N-terminal residues 1-173 of Venus (designated VN-173) to produce SIRT1-Venus N-terminus-encoding plasmid (SIRT1-VN). CLOCK-C-terminal of Venus (VC), BMAL1-VC, and BMAL1 deletion mutants were previously described [22].
NIH3T3, HeLa, and COS7 cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM, Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Invitrogen). Cells were cultured at 37℃ in a humidified 5% CO2 environment. For transient transfections, cells were seeded at a density of 1×105 cells per well in a 12-well plate. Cells were then transfected using Lipofectamine PLUS (Invitrogen) or Metafectene EASY (Biontex, San Diego, CA, USA) reagents according to the manufacturer's protocol.
Cell extracts were prepared from HeLa cells transfected with the plasmids indicated. Proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (6% polyacrylamide) and then electrophoretically transferred onto a polyvinylidene difluoride membrane (Immobilon P, Millipore, Billerica, MA, USA). Target proteins were detected with anti-N-term green fluorescent protein (GFP, Sigma Aldrich, St. Louis, MO, USA). The immunoreactive bands were visualized with an enhanced chemiluminescent detection kit (Thermo Fisher Scientific, Rockford, IL, USA).
Details of the BiFC protocol have been described previously [22]. Briefly, COS7 cells were transfected with various BiFC expression vectors. Twelve hours after transfection, cells were fixed with 3.7% paraformaldehyde and washed twice with ice-cold phosphate-buffered saline (PBS). After fixation, cells were stained with 4',6-diamidino-2-phenylindole (DAPI) diluted in mounting solution and mounted onto glass slides. To capture BiFC images, yellow fluorescent protein (YFP) excitation and emission filters (EM) were used (excitation filter=500 nm, dichroic mirror=515 nm, EM=535 nm).
For immunostaining procedures, 1×105 COS7 cells were seeded per well onto coated cover glass in 12-well cell culture plates. The following day, cells were transfected with 200, 100, and 100 ng of SIRT1-VN, CLOCK-VC, and BMAL1-VC (including deletion mutants) plasmids, respectively. Twelve hours after transfection, cells were fixed with 3.7% paraformaldehyde for 15 minutes. Cell membranes were then permeabilized with 0.5% Triton X-100 in PBS for 5 minutes. Cells were then blocked with donkey serum for 30 minutes. Target proteins were detected with anti-N-term GFP (Sigma Aldrich), anti-CLOCK (Santa Cruz Biotechnologies, Santa Cruz, CA, USA), and anti-BMAL1 antibodies [23]. Primary antibodies were diluted 1:200 in blocking solution (donkey serum) and applied for 1 hour. The secondary antibody was diluted 1:100 in blocking solution and applied for no longer than 1 hour. After antibody incubations, cells were washed with 0.1% Triton X-100 in PBS, and cover slips were transferred onto slides. For DAPI staining, the DAPI solution was diluted in mounting media. Cells were imaged by fluorescence microscopy.
NIH3T3 cells were seeded at a density of 5×104 cells per well onto a 24-well plate and transfected with reporter plasmids Ebox-Luc (10 ng per well) [24], Per1-Luc (10 ng per well), and pRL-TK (50 ng per well) in combination with or without effector genes (CLOCK, BMAL1, and SIRT1: 200 ng per well each). The total amount of DNA used was held constant by adding the empty pcDNA3.1 plasmid. Twenty-four hours after transfection, cells were incubated in 0, 10, or 100 µM resveratrol for another 24 hours. Cell extracts were then incubated in 0.3 mL passive lysis buffer (Promega, Madison, WI, USA) for 15 minutes at room temperature. Luciferase activities were measured with a commercial Dual Luciferase assay kit (Promega).
Statistical tests were carried out using Sigma Plot version 8.0 (Systat Software, San Jose, CA, USA). Values are expressed as mean±SEM of independent experiments. For comparison of three or more groups, data were analyzed by one-way analysis of variance followed by post hoc Tukey's multiple comparison test or Dunnett's multiple comparison test. A value of P less than 0.05 was considered statistically significant.
Previous studies have shown that endogenous SIRT1 interacts with both CLOCK and BMAL1. To investigate this interaction in intact cells, we conducted a BiFC analysis that visualized the direct interaction between SIRT1 and CLOCK or BMAL1 under native cell conditions. SIRT1-VN was transfected into COS7 cells, and its expression was verified by Western blot analysis (Fig. 1A, B). Subsequently, cells were expressed SIRT1-VN with CLOCK-VC or BMAL1-VC, and the BiFC signal of the cells was imaged. As shown in Fig. 1C, a positive BiFC signal was observed in most cell nuclei, suggesting a direct interaction between SIRT1 and both CLOCK-VC and BMAL1-VC. Immunocytochemistry, performed using both anti-CLOCK and -BMAL1 antibodies, showed that cytosolic CLOCK proteins detected in the nucleus colocalized with BiFC signals. The nuclear presence of these proteins suggests that the nuclear localization sequence (NLS) of SIRT1 is responsible for its nuclear localization (Fig. 1C; second lane from the top, middle panel).
To further explore SIRT1-BMAL1 interactions, we delineated which domain of BMAL1 is critical for binding to SIRT1. Fig. 2A shows a schematic diagram of mouse BMAL1, depicting several important domains, including a NLS, DNA-binding and protein interaction motifs (bHLH, PAS-A, and PAS-B), and a transcriptional activation domain. To test whether any of these domains are necessary for the SIRT1-BMAL1 interaction, we conducted BiFC analysis using region-specific deletion mutants corresponding to the aforementioned domains. COS7 cells were cotransfected with the SIRT1-VN expression vector and each of the various BMAL1-VC deletion mutant expression vectors. The BMAL1 deletion mutants all yielded BiFC signals that were comparable to wild-type BMAL1. However, deletion of the PAS-B domain substantially weakened BiFC signal intensity, suggesting reduced binding to SIRT1 (Fig. 2B). Immunocytochemical analysis using anti-BMAL1 antibody indicated that the attenuated intensity of the BiFC signal was not caused by the weak expression of BMAL1.
We hypothesized that resveratrol, a SIRT1 activator, not only enhances the activity of SIRT1, but can also influence its interaction with the CLOCK/BMAL1 complex. To test this hypothesis, BiFC analysis was performed with COS7 cells co-expressing SIRT1-VN and either CLOCK-VC or BMAL1-VC. Cells were then exposed to either resveratrol or control vehicle prior to BiFC analysis. The addition of resveratrol did not induce any significant change in the BiFC signal, suggesting that the interaction of SIRT1 with either CLOCK or BMAL1 was not significantly affected (Fig. 3A). For a quantitative analysis of BiFC data, ten microscope fields of view were randomly selected, and BiFC-positive signals were divided by DAPI signals (Fig. 3B). Taken together, our data suggest that, although resveratrol affects SIRT1 activity, it does not modulate the interaction of SIRT1 with the CLOCK/BMAL1 complex.
Although we did not find that resveratrol produced any significant effects on the interaction of SIRT1 with CLOCK or BMAL1, we further investigated how resveratrol affects SIRT1 regulation of clock gene transcription. Luciferase assays were performed using two reporters: an E-box-reporter construct (Ebox-Luc) and a full-length Per1-Luciferase reporter (Per1-Luc). CLOCK and BMAL1 expression vectors were cotransfected into NIH3T3 cells along with each reporter construct. Administration of resveratrol (10 or 100 µM) for 24 hours gradually decreased Ebox-Luc transcription activity (Fig. 4A). Resveratrol (100 µM) also significantly reduced Per1-Luc transcription activity under basal as well as CLOCK/BMAL1-induced conditions (Fig. 4B). These findings suggest that resveratrol likely activates endogenous SIRT1, leading to the repression of clock gene expression (in this instance, Per1).
To determine the combined effects of SIRT1 and resveratrol on CLOCK/BMAL1-mediated transcription of Per1, we treated cells coexpressing SIRT1, CLOCK, and BMAL1 with increasing concentrations of resveratrol (10 and 100 µM). Exposure of cells to 100 µM resveratrol for 24 hours significantly repressed the basal and CLOCK/BMAL1-induced promoter activity when SIRT1 was coexpressed (Fig. 4C, D).
It has been reported that endogenous SIRT1 interacts with CLOCK and/or BMAL1 [9,10]. These interactions, observed primarily using biochemical approaches, remain controversial. Immunoprecipitation assays showed that ectopically expressed BMAL1 did not coimmunoprecipitate with the SIRT1-CLOCK complex [9]. Other studies have shown that SIRT1 can interact with BMAL1 in transfected cells and mouse liver tissues [10,12]. The discrepancies in these studies may result from either the different experimental conditions used in the immunoprecipitation assays or undiscovered variations among the expression plasmids used. Thus, in this study we attempted to visualize the SIRT1-CLOCK/BMAL1 interaction using BiFC imaging in cells. Accordingly, this technique enabled the detection of SIRT1 interaction with the CLOCK/BMAL1 complex in a native cellular environment. Moreover, we used BiFC imaging to further analyze several BMAL1 domains for their roles in SIRT1 interaction. We found that a BMAL1 construct lacking the PAS-B domain produced fewer BiFC-positive signals (Fig. 2). Together with a gradually increasing quantity of structural information on the CLOCK/BMAL1 complex [25], these SIRT1-BMAL1 interaction data can provide valuable insight to elucidate the functional relevance of SIRT1 action in the circadian clockwork.
Resveratrol treatment had little effect on the interaction of SIRT1 with CLOCK/BMAL1, as revealed by BiFC assay results (Fig. 3). These data coincide with results from a previous study, in which the SIRT1-CLOCK interaction was not significantly affected by the treatment of various effectors of SIRT1 function [9]. However, transcriptional assays using the Per1 promoter and E-box-based luciferase activity showed that SIRT1 activation via treatment with resveratrol significantly downregulated CLOCK/BMAL1-mediated transcriptional activation (Fig. 4).
Indeed, there are discrepancies with regard to the involvement of SIRT1 in the circadian clock mechanism. Nakahata et al. [9] reported that SIRT1 contributes to the mRNA expression of circadian genes by acting as a negative regulator and controls the acetylation/deacetylation status of BMAL1 through CLOCK. Asher et al. [10] suggested that SIRT1 deacetylates PER2 and consequently influences the high levels of circadian gene expression. The repressive role of SIRT1 in circadian clock machinery was further supported by results from Ramsey et al. [12], which suggest that NAD+ functions as a metabolic oscillator and negatively regulates core clock gene expression via SIRT1. Consistent with these results, our promoter assays showed that SIRT1 inhibits CLOCK/BMAL1-mediated transcriptional activity on the Per1 promoter, indicating a negative role in circadian clock gene expression. In addition, resveratrol treatment potentiated the inhibitory effect of SIRT1 on CLOCK/BMAL1-mediated transcription. Therefore, we conclude that SIRT1 and resveratrol synergistically repress circadian gene expression through the enhancement of deacetylase activity.
In this study, however, we did not explore the mechanism of action behind resveratrol-induced SIRT1 deacetylase activity; thus, further studies are needed to elucidate this biochemical interaction. Since resveratrol has various target molecules, an experimental approach such as an in vitro deacetylation assay targeting SIRT1 with its substrate molecules will be necessary to assess whether resveratrol increases SIRT1 deacetylase activity. Moreover, it is of interest to examine whether resveratrol inhibits the endogenous expression of clock genes at both mRNA and/or protein levels using quantitative reverse transcription-PCR and Western blot analysis, respectively.
Transcriptome profiling studies have shown that many genes related to metabolism are rhythmically expressed [26,27]. The dominance of the feeding cycle as a zeitgeber for peripheral clocks implies that the circadian clock plays an important role in nutrient processing and energy homeostasis [28]. In this regard, SIRT1 would be an important regulator of circadian clockwork by serving as a molecular linker between circadian clock and metabolism with regard to epigenetic regulation [29]. It has been reported that SIRT1 helps cells resist oxidative or radiation-induced stresses [16,30] as well as promotes fat mobilization from white adipose tissue, events that contribute to extending life span [31]. It would be valuable to study these multimodal functions of SIRT1 from the perspective of circadian regulation.
In summary, we demonstrate that SIRT1 forms a complex with CLOCK/BMAL1 in the cell nucleus. Additionally, our data suggest a role for SIRT1 as a negative regulator of circadian gene expression, likely via its deacetylase activity.
Figures and Tables
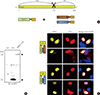 | Fig. 1SIRTUIN1 interacts with CLOCK and BMAL1 in cell nuclei. (A) Schematic diagram of Venus-based bimolecular fluorescence complementation (BiFC) constructs. (B) Expression of SIRT1-N-terminal of Venus (VN) was detected by sodium dodecyl sulfate polyacrylamide gel electrophoresis separation followed by immunoblotting with an anti-green fluorescent protein (GFP) antibody (arrows). (C) Results from BiFC analysis and immunocytochemical experiments (ICC). COS7 cells were coexpressed with SIRT1-VN and either CLOCK-C-terminal of Venus (VC) or BMAL1-VC. Cells were immunostained with anti-GFP, anti-CLOCK, and anti-BMAL1 antibodies (red) to detect SIRT1-VN, CLOCK-VC, and BMAL1-VC, respectively. Nuclei were visualized with 4',6-diamidino-2-phenylindole (DAPI; blue). The images were acquired by fluorescence microscopy using specific filter sets for yellow fluorescent protein and red fluorescent protein. Scale bar=10 µm. |
 | Fig. 2Identification of the SIRTUIN1 (SIRT1)-binding domain of BMAL1. (A) Schematic diagram of mouse BMAL1. (B) Binding of SIRT1 to BMAL1 deletion mutants was analyzed by bimolecular fluorescence complementation (BiFC). COS7 cells were transfected with plasmids encoding SIRT1-N-terminal of Venus (VN) and BMAL1-C-terminal of Venus (VC) wildtype (WT) or its various mutants (Δnuclear localization sequence [NLS], BMAL1 without a functional nuclear localization signal; Δbasic-helix-loop-helix [bHLH], encoding BMAL1 Δ71 to 140; ΔPer-Arnt-Sim [PAS]-A, BMAL1 Δ210 to 320; ΔPAS-B, BMAL1 Δ350 to 480; Δtranscriptional activation domain [TAD], BMAL1 Δ553 to 625). The images were captured by fluorescence imaging microscopy using specific filter sets for yellow fluorescent protein and red fluorescent protein. Scale bar=10 µm. ICC, immunocytochemical; DAPI, 4',6-diamidino-2-phenylindole. |
 | Fig. 3Interaction of SIRTUIN1 (SIRT1) with CLOCK and BMAL1 upon resveratrol treatment. (A) COS7 cells were transfected with expression plasmids encoding SIRT1-N-terminal of Venus (VN) and either CLOCK-C-terminal of Venus (VC) or BMAL1-VC. Twelve hours after transfection, cells were treated with 100 µM resveratrol for 6 hours and then imaged. Scale bar=20 µm. (B) Bimolecular fluorescence complementation (BiFC) signals from control and resveratrol-stimulated groups were compared with signals from 4',6-diamidino-2-phenylindole (DAPI)-stained cells. Values are expressed as mean±SEM % (n=10 fields for each group). Veh, vehicle; RSV, resveratrol; YFP, yellow fluorescent protein. |
 | Fig. 4The effects of resveratrol on Ebox-Luc and Per1-Luc transcriptional activities. NIH3T3 cells expressing wild-type CLOCK, BMAL1, and either (A) Ebox-Luc or (B) Per1-Luc promoters were treated with increasing amounts of resveratrol (10 and 100 µM) to determine transcriptional activity. Luciferase activities in cell extracts were analyzed and normalized by cotransfected pRL-TK activity in each sample. To examine the effects of SIRTUIN1 (SIRT1) and resveratrol on the transcriptional activity of (C) Ebox-Luc and (D) Per1-Luc, SIRT1 was expressed with or without CLOCK and BMAL1 in NIH3T3 cells and incubated in resveratrol (10 and 100 µM). Values are expressed as mean±SEM in arbitrary units (AU), where the mean activity of the empty vector was set at 1 (n=3). aP<0.01 vs. vehicle-treated group. |
ACKNOWLEDGMENTS
We thank Dr. Gad Asher (University of Geneva, Geneva, Switzerland) for providing the HA-FLAG-tagged human SIRT1 clone. This work was supported by grants from the Korea Ministry of Education, Science, and Technology (MEST) through the Brain Research Center of the 21st Century Frontier Research Program (2009K001287) and the BK21 Plus program through the National Research Foundation of Korea funded by the Ministry of Education (10Z20130012420). BioScience Writers edited the manuscript.
References
1. Son GH, Chung S, Kim K. The adrenal peripheral clock: glucocorticoid and the circadian timing system. Front Neuroendocrinol. 2011; 32:451–465.
2. Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010; 72:517–549.
3. Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008; 9:764–775.
4. Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002; 110:251–260.
5. Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004; 43:527–537.
6. Kaasik K, Lee CC. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature. 2004; 430:467–471.
7. Rutter J, Reick M, McKnight SL. Metabolism and the control of circadian rhythms. Annu Rev Biochem. 2002; 71:307–331.
8. Tu BP, McKnight SL. Metabolic cycles as an underlying basis of biological oscillations. Nat Rev Mol Cell Biol. 2006; 7:696–701.
9. Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008; 134:329–340.
10. Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008; 134:317–328.
11. Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009; 324:654–657.
12. Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S, Bass J. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009; 324:651–654.
13. Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004; 73:417–435.
14. Dali-Youcef N, Lagouge M, Froelich S, Koehl C, Schoonjans K, Auwerx J. Sirtuins: the 'magnificent seven', function, metabolism and longevity. Ann Med. 2007; 39:335–345.
15. Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A, Easlon EJ, Lin SJ, Guarente L. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006; 4:e31.
16. Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004; 303:2011–2015.
17. Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004; 116:551–563.
18. Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003; 425:191–196.
19. Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006; 127:1109–1122.
20. Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006; 444:337–342.
21. Vetterli L, Maechler P. Resveratrol-activated SIRT1 in liver and pancreatic beta-cells: a Janus head looking to the same direction of metabolic homeostasis. Aging (Albany NY). 2011; 3:444–449.
22. Lee Y, Lee J, Kwon I, Nakajima Y, Ohmiya Y, Son GH, Lee KH, Kim K. Coactivation of the CLOCK-BMAL1 complex by CBP mediates resetting of the circadian clock. J Cell Sci. 2010; 123(Pt 20):3547–3557.
23. Kwon I, Lee J, Chang SH, Jung NC, Lee BJ, Son GH, Kim K, Lee KH. BMAL1 shuttling controls transactivation and degradation of the CLOCK/BMAL1 heterodimer. Mol Cell Biol. 2006; 26:7318–7330.
24. Son GH, Chung S, Choe HK, Kim HD, Baik SM, Lee H, Lee HW, Choi S, Sun W, Kim H, Cho S, Lee KH, Kim K. Adrenal peripheral clock controls the autonomous circadian rhythm of glucocorticoid by causing rhythmic steroid production. Proc Natl Acad Sci U S A. 2008; 105:20970–20975.
25. Huang N, Chelliah Y, Shan Y, Taylor CA, Yoo SH, Partch C, Green CB, Zhang H, Takahashi JS. Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex. Science. 2012; 337:189–194.
26. Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007; 5:e34.
27. Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002; 109:307–320.
28. Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010; 330:1349–1354.
29. Masri S, Sassone-Corsi P. The circadian clock: a framework linking metabolism, epigenetics and neuronal function. Nat Rev Neurosci. 2013; 14:69–75.
30. Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001; 107:137–148.
31. Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004; 429:771–776.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download