Abstract
Obesity gives vent to many diseases such as type 2 diabetes, hypertension, and hyperlipidemia, being considered as the main causes of mortality and morbidity worldwide. The pathogenesis and pathophysiology of metabolic syndrome can well be understood by studying the molecular mechanisms that control the development and function of adipose tissue. In human body, exist two types of adipose tissue, the white and the brown one, which are reported to play various roles in energy homeostasis. The major and most efficient storage of energy occurs in the form of triglycerides in white adipose tissue while brown adipose tissue actively participates in both basal and inducible energy consumption in the form of thermogenesis. Recent years have observed a rapid and greater interest towards developmental plasticity and therapeutic potential of stromal cells those isolated from adipose tissue. The adipocyte differentiation involves a couple of regulators in the white or brown adipogenesis. Peroxisome proliferators-activated receptor-γ actively participates in regulating carbohydrate and lipid metabolism, and also acts as main regulator of both white and brown adipogenesis. This review based on our recent research, seeks to highlight the adipocyte differentiation.
Obesity has emerged as one of the deadliest life threat of the 21st century with more than one billion adults classified as an overweight. Obesity is most likely to cause diseases such as type 2 diabetes, hypertension, hyperlipidemia, which are considered as the leading causes of mortality and morbidity globally. It has been reported by a number of researchers that mesenchymal cell within the stromal-vascular fraction of subcutaneous adipose tissue displays multilineage developmental plasticity. This review has been focused towards the differentiation of both the brown adipose tissue (BAT) as well as the white one presenting the recent evidence with our results.
Adipose tissue is composed of numerous mesenchymal stem cells (MSCs) which differentiate into multiple lineages such as adipocytes in vitro, providing a unique platform to study molecular machineries of adipocyte differentiation. The fat progenitors are quite easy to yield and can be found in plenty in the stromal vascular fraction, which is isolated through collagenase dissociation of the fat tissue and size-filtration of the constituent cells which have alternatively been referred to as processed lipoaspirate cells, adipose-derived stem cells, adipose-derived stromal cells, and adipose-derived mesenchymal progenitor cells. The cells previously considered preadipocytes may essentially the same in population. These cells known as adipose tissue-derived stromal cells (ADSC), may clearly explain their origin and character [1]. ADSC and MSCs are found to be quite similar as they both represent the stromal cell fraction isolated from an adipose depot (subcutaneous tissue for the former, bone marrow for the latter) on the basis of adherence to tissue culture plastic. While an extensive research has been devoted to the phenotypic characterization of MSCs, the one that's of ADSC is still in its initial stages. Our analysis of stem cell-related surface markers, reported the presence of Sca1(ly-6A/E) and CD44 antigen but did not exhibit the presence of c-Kit, Lin, CD11b, CD31, CD34, or CD45 in ADSC obtained from C57Bl/6 mice [1]. In an earlier research, MSCs obtained from C57Bl/6 mice were reported to have a high expression of Sca1, CD34, but not that's of c-kit, Lin, CD11b, CD31, CD45 [2]. This shows ADSC to have a similar population of MSCs, but not that's of hematopoietic stem cells obtained from bone marrow. Importantly, these characteristics of ADSC surface markers were conserved after several passages of cell expansion.
Since a number of cytokines are reported to be secreted from adipose tissue, the secretion of angiogenesis-related cytokines from ADSC has highly been investigated. The analysis of quantitative real time polymerase chain reaction, reported the relatively high expression of hepatocyte growth factor (HGF), vascular endothelial growth factor (VEGF), placentral growth factor, and transforming growth factor-β, the moderate expression of fibroblast growth factor-2 and angiopoietin (Ang)-1, and the low level of Ang-2 from ADSC, and ADSC was also reported to secret multiple angiogenic growth factors, such as VEGF and HGF, at levels that are bioactive [3,4]. Microarrays have recently been used by Katz et al. [5] to evaluate the genes relating to angiogenesis and extracellular matrix in undifferentiated human ADSC isolated from three separate donors. The most highly transcribed genes were found to be related to functional groupings such as cell adhesion, matrix proteins, growth factors and receptors, and proteases, and the transcriptome of ADSC revealed many similarities to the profiles of bone marrow MSCs. Importantly, the recent work suggests that attachment of ADSC to culture plastic and their duration in culture (i.e., time on plastic) and the culture medium used significantly change the cell surface phenotypic profile of each cells.
Adipocyte differentiation comprises a plethora of transcription factors. The proliferator-activated receptor gamma (PPARγ) and members of the CCAAT/enhancer binding protein (C/EBP) family of transcription factors [6,7] are considered as the important ones and play vital roles. When the adipogenic stimulation is complete, the expressions of C/EBPδ and C/EBPβ are induced in early phase, and the ones of C/EBPα and PPARγ2 are expressed later by transcriptional regulation [7,8]. These two transcription factors, C/EBPα and PPARγ2, might be central to the control of adipocyte-specific gene expression. Critical aspects of the transcriptional control of adipocyte differentiation still need to be cleared. For instance, although C/EBPβ/δ double knockout mice exhibit a remarkably small mass of epididymal WAT, levels of PPARγ and C/EBPα expression are similar to those in wild-type mice [8], indicating that other factors are also involved in the transcriptional control of adipocyte differentiation. In fact, the involvement of a number of additional transcription factors have been reported to function in adipogenesis, such as the ones Wnt, Foxo1, CREB, ADD1/SREBP-1c, and members of the Krüppel-like factor (KLF) family of transcription factors (i.e., KLF5, 15) [9-11].
BAT has been reported to actively participate both in basal and in inducible energy expenditure in the form of thermogenesis. It is formed through expression of uncoupling protein-1 (UCP1), a 32 kDa protein found in the inner mitochondrial membrane that allows dissipation of the proton electrochemical gradient generated by respiration in the form of heat [12]. In human, BAT is found to be present in fetuses and newborns at axillary, cervical, perirenal, and periadrenal regions but soon after birth it starts vanishing and traditionally becomes extinct in adults. Recent morphological and scanning studies have shown that brown fat in humans may not be as rare as once believed. In fact, the areas of metabolically active brown fat in the cervical, supraclavicular, axillary, and paravertebral regions of normal individuals can be detected by 18F-fludeoxyglucose positron emission tomography [13-16].
In human, BAT exists in neonates where it combusts the energy to generate heat, instead of storing. The processes are executed in mitochondria, by the molecules such as UCP1 through the process called energy uncoupling. The presence of BAT has recently been reported in adult human. The higher activity presented by them in the participants subjected to cold environment, is enough to validate them physiologically functional. Understanding the technique of regulating the generation of BAT carries a great therapeutic impact, because energy consumption in BAT is reported to bring a decline in the lipid storage in WAT. An increase in brown adipocytes can be brought by cold or β3-adrenergic stimulation. The WAT-derived progenitor cells have recently been reported to undergo brown adipogenesis in vitro in both mice [17,18] and humans [17,19], but their molecular mechanism is still unclear. C/EBPβ [20-23], along with PRDM16 [20,24], induces the brown fat program demonstrating its essential role in brown adipogenesis. Transcription factors are found to induce or suppress the transcription of target genes through coordination with chromatin remodeling factors. Differentiation can be calculated by the coordinating roles of transcription factors and chromatin remodeling factors, and to know the technique to regulate the entire process is an intriguing topic considering the therapeutic potential of, for example, histone deacetylases (HDAC) inhibitor, already widely used in various kinds of disease models with proven efficacy [25].
The homeotic genes as a novel regulator of adipogenesis have been focused in our research. Homeotic genes are characterized as the representative example of developmental genes, and help to determine the anterior-posterior axis of the body plan in the early phases of morphogenesis. However, their roles after development have not been full defined. Since the developmental process is regulated by Hox genes, Hox genes are also likely to regulate the differentiation or commitment of tissue progenitors postnatally. Regarding this, Hox genes engage in the regulation of tissue stem cells. HoxA11 suppresses the myoblast differentiation by suppressing the expression of MyoD, a master regulator of myogenesis, which is regulated by miR-181 [26]. HoxB5b is reported to restrict the number of the cardiac atrial cells, thereby limiting the heart chamber size [27]. The homeobox gene HoxA9 is usually found to be present in primitive bone marrow cells, and overexpression of HoxA9 is reported to markedly expand hematopoietic stem cells [28].
A characteristic difference is reported between BAT and WAT expression patterns of the Hox family of homeobox genes (Hox genes) [29-31]. Among the 39 Hox genes, only HOXC6 and HOXC8 genes were reported to be present extensively in human ADSCs. However the expression of HOXC6 was found to be relatively broad, the expression of HOXC8 was highly enriched in WAT and BAT. Interestingly, overexpressed HOXC8 was reported to completely suppress adipogenesis in ADSCs. After adipocyte differentiation, the protein expression of HOXC8 disappeared in the differentiated adipocytes, however the significant decrease in the HOXC8 mRNA was not observed. This result confirms the efficacy of posttranscriptional manner for regulating the expression of HOXC8.
We addressed to microRNA (miRNA), which facilitates post-transcriptional regulation. The miRNAs are reported as small, nonprotein-coding RNAs that are paired with specific mRNAs and inhibit translation or promote mRNA degradation [32]. The genes encoding miR-196a are located upstream of Hox9 paralogs in the B and C clusters [33]. The miR-196a was reported to suppress the expression of HOXC8 posttranscriptionally without degrading mRNA [34]. In our study, the expression of miR-196a was induced along adipogenesis in ADSCs for approximately 5.0 fold. In rodents, β3-adrenaline agonist or exposure to cold environment induces ectopic brown adipocytes in WAT [24]. Cyclic adenosine monophosphate (cAMP) signaling also actively participates in brown adipogenesis, in fact, miR-196a is induced by cAMP cultivated by adipogenic induction media or physiological stimulation such as cold environment and β3 adrenaline stimulation and plays a significant role in adipogenesis.
A microarray analysis in HOXC8-transduced ADSCs was performed for analyzing the molecular targets of HOXC8. Although PPARγ, aP2, and C/EBPα, marker genes for white adipocytes, were upregulated in differentiated ADSCs, a series of brown adipocyte signature genes, such as C/EBPβ, PGC-1α, and UCPs were also upregulated. These results implied that even if ADSCs were derived from human flank fat mainly comprising white adipocytes, a molecular network for brown adipogenesis was also driven in ADSCs differentiated in vitro. Interestingly, HOXC8 was found to suppress a number of adipogenesis-related genes of white and brown adipocyte.
In the analysis of chromatin immunoprecipitation assay, significant binding was suggested in the 3'UTR sequence of C/EBPβ gene. Transcription factors have been reported to regulate the transcription through binding to 3'UTR of the gene. Therefore, the 3'UTR sequence of C/EBPβ gene was cloned and inserted into the 3' end of luciferase gene. Only when inserted in the oriented direction, C/EBPβ 3'UTR showed transcriptional activity by the luciferase activity, validating C/EBPβ 3'UTR as a functional region for transcription. Furthermore, cotransfection of HOXC8 expression vector was found to suppress the luciferase expression driven by C/EBPb 3'UTR whereas HOXC8 exhibited no suppressive effect with mutated homeodomain in which four amino acids essential for DNA recognition were substituted for alanine.
We generated transgenic mice expressing miR-196a under the control of the aP2 (also called FABP4) promoter/enhancer that is found to be active exclusively in fat tissues [34]. The transgenic mice (hereafter, miR-196a mice) were produced at a Mendelian ratio and were viable. The miR-196a mice were observed to gain weight quite slowly as compared to WT mice. More detailed inspections revealed a low quantity of inguinal fat, epididymal fat and liver in miR-196a mice despite that the weight of heart and soleus muscle were not significantly changed, which indicates that the decreased body weight was due to reduced fat accumulation.
Histological analysis revealed that inguinal fat of miR-196a mice comprised smaller adipocytes as compared to that of WT. Furthermore, the inguinal fat of miR-196a mice exhibited interspersed multiple regions with clustered multiocular cells with the expression of UCP1 indicative of brown adipocytes. We focused our attention to find whether these ectopic brown adipocyte-like cells were metabolically active or not by measuring the metabolic status of miR-196a mice. In indirect calorimetry miR-196a mice as compared to those of WT were found to consume higher amount of oxygen, both in the light and dark phases of a day. Energy expenditure, calculated on the basis of oxygen and carbon dioxide consumption, was found to have been maximized in miR-196a mice. According to results, the brown adipocyte-like cells generated in WAT were found to promote the energy consumption, and likely to be detected in individual animals.
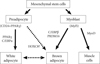
In summary, our results attributed a new role to Hox genes in regulation of fat progenitors. The expression of HOXC8 was regulated by miR-196a induced by various kinds of adipogenic stimuli converging to cAMP signaling. HOXC8 was reported to suppress C/EBPβ expression by recruiting HDAC to 3'UTR regulatory site of the C/EBPβ gene. Modulation of this pathway by miR-196a in mice induced higher expression of C/EBPβ-responsive network in fat tissue and ectopic brown adipocyte-like cells rendering the mice resistant to obesity (Fig. 1).
Our research came up with one example in which increased brown adipocyte-like cells in WAT were found to resist obesity with favorable glucose metabolism. The increased brown adipocytes in WAT were metabolically functional, because of higher energy expenditure, resistance to obesity and improved glucose metabolism exhibited by the mice. Further elucidation of the molecular mechanisms that control adipogenesis would help to introduce new molecular targets for therapeutics of intractable obesity and related diseases.
Figures and Tables
Fig. 1
Adipocyte and myocyte differentiation. Characterization of regulatory regions of adipose-specific genes has helped in the discovery of the transcription factors peroxisome proliferator-activated receptor gamma (PPARγ) and CCAAT/enhancer binding protein (C/EBP), which play a key role in the complex transcriptional cascade during white adipocyte differentiation. C/EBPβ, in collaboration with PR domain containing 16 (PRDM16), induces the brown fat program demonstrating its essential role in brown adipogenesis, which was partially originated from myoblast. Although, preadipocytes have been found likely to undergo brown adipogenesis, the molecular mechanism still needs to be clarified. This study reports HOXC8 as a novel candidate.
ACKNOWLEDGMENTS
This work has partially been funded by the Ministry of Education, Culture, Sports, Science and Technology; the Japan Heart Association.
References
1. Nakagami H, Maeda K, Morishita R, Iguchi S, Nishikawa T, Takami Y, Kikuchi Y, Saito Y, Tamai K, Ogihara T, Kaneda Y. Novel autologous cell therapy in ischemic limb disease through growth factor secretion by cultured adipose tissue-derived stromal cells. Arterioscler Thromb Vasc Biol. 2005. 25:2542–2547.
2. Peister A, Mellad JA, Larson BL, Hall BM, Gibson LF, Prockop DJ. Adult stem cells from bone marrow (MSCs) isolated from different strains of inbred mice vary in surface epitopes, rates of proliferation, and differentiation potential. Blood. 2004. 103:1662–1668.
3. Rehman J, Considine RV, Bovenkerk JE, Li J, Slavens CA, Jones RM, March KL. Obesity is associated with increased levels of circulating hepatocyte growth factor. J Am Coll Cardiol. 2003. 41:1408–1413.
4. Rehman J, Traktuev D, Li J, Merfeld-Clauss S, Temm-Grove CJ, Bovenkerk JE, Pell CL, Johnstone BH, Considine RV, March KL. Secretion of angiogenic and antiapoptotic factors by human adipose stromal cells. Circulation. 2004. 109:1292–1298.
5. Katz AJ, Tholpady A, Tholpady SS, Shang H, Ogle RC. Cell surface and transcriptional characterization of human adipose-derived adherent stromal (hADAS) cells. Stem Cells. 2005. 23:412–423.
6. Morrison RF, Farmer SR. Hormonal signaling and transcriptional control of adipocyte differentiation. J Nutr. 2000. 130:3116S–3121S.
7. Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev. 2000. 14:1293–1307.
8. Tanaka T, Yoshida N, Kishimoto T, Akira S. Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene. EMBO J. 1997. 16:7432–7443.
9. Nakae J, Kitamura T, Kitamura Y, Biggs WH 3rd, Arden KC, Accili D. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev Cell. 2003. 4:119–129.
10. Oishi Y, Manabe I, Tobe K, Tsushima K, Shindo T, Fujiu K, Nishimura G, Maemura K, Yamauchi T, Kubota N, Suzuki R, Kitamura T, Akira S, Kadowaki T, Nagai R. Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 2005. 1:27–39.
11. Gray S, Feinberg MW, Hull S, Kuo CT, Watanabe M, Sen-Banerjee S, DePina A, Haspel R, Jain MK. The Kruppel-like factor KLF15 regulates the insulin-sensitive glucose transporter GLUT4. J Biol Chem. 2002. 277:34322–34328.
12. Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004. 84:277–359.
13. Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. Am J Physiol Endocrinol Metab. 2007. 293:E444–E452.
14. Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, Kolodny GM, Kahn CR. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009. 360:1509–1517.
15. van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJ. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009. 360:1500–1508.
16. Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto NJ, Enerback S, Nuutila P. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009. 360:1518–1525.
17. Schulz TJ, Huang TL, Tran TT, Zhang H, Townsend KL, Shadrach JL, Cerletti M, McDougall LE, Giorgadze N, Tchkonia T, Schrier D, Falb D, Kirkland JL, Wagers AJ, Tseng YH. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc Natl Acad Sci U S A. 2011. 108:143–148.
18. Petrovic N, Walden TB, Shabalina IG, Timmons JA, Cannon B, Nedergaard J. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J Biol Chem. 2010. 285:7153–7164.
19. Elabd C, Chiellini C, Carmona M, Galitzky J, Cochet O, Petersen R, Penicaud L, Kristiansen K, Bouloumie A, Casteilla L, Dani C, Ailhaud G, Amri EZ. Human multipotent adipose-derived stem cells differentiate into functional brown adipocytes. Stem Cells. 2009. 27:2753–2760.
20. Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, Spiegelman BM. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009. 460:1154–1158.
21. Wu Z, Xie Y, Bucher NL, Farmer SR. Conditional ectopic expression of C/EBP beta in NIH-3T3 cells induces PPAR gamma and stimulates adipogenesis. Genes Dev. 1995. 9:2350–2363.
22. Karamanlidis G, Karamitri A, Docherty K, Hazlerigg DG, Lomax MA. C/EBPbeta reprograms white 3T3-L1 preadipocytes to a Brown adipocyte pattern of gene expression. J Biol Chem. 2007. 282:24660–24669.
23. Kajimura S, Seale P, Spiegelman BM. Transcriptional control of brown fat development. Cell Metab. 2010. 11:257–262.
24. Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, Cohen P, Cinti S, Spiegelman BM. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest. 2011. 121:96–105.
25. Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009. 10:32–42.
26. Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, Cuvellier S, Harel-Bellan A. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006. 8:278–284.
27. Waxman JS, Keegan BR, Roberts RW, Poss KD, Yelon D. Hoxb5b acts downstream of retinoic acid signaling in the forelimb field to restrict heart field potential in zebrafish. Dev Cell. 2008. 15:923–934.
28. Lawrence HJ, Christensen J, Fong S, Hu YL, Weissman I, Sauvageau G, Humphries RK, Largman C. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 2005. 106:3988–3994.
29. Cantile M, Procino A, D'Armiento M, Cindolo L, Cillo C. HOX gene network is involved in the transcriptional regulation of in vivo human adipogenesis. J Cell Physiol. 2003. 194:225–236.
30. Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007. 131:242–256.
31. Timmons JA, Wennmalm K, Larsson O, Walden TB, Lassmann T, Petrovic N, Hamilton DL, Gimeno RE, Wahlestedt C, Baar K, Nedergaard J, Cannon B. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc Natl Acad Sci U S A. 2007. 104:4401–4406.
32. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009. 136:215–233.
33. Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004. 304:594–596.
34. Mori M, Nakagami H, Rodriguez-Araujo G, Nimura K, Kaneda Y. Essential role for miR-196a in brown adipogenesis of white fat progenitor cells. PLoS Biol. 2012. 10:e1001314.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download