Abstract
Type 2 diabetes mellitus (T2DM) is caused by complex interplay between multiple genetic and environmental factors. The three major approaches used to identify the genetic susceptibility include candidate gene approach, familial linkage analysis and genome- wide association analysis. Recent advance in genome-wide association studies have greatly improved our understanding of the pathophysiology of T2DM. As of the end of 2010, there are more than 40 confirmed T2DM-associated genetic loci. Most of the T2DM susceptibility genes were implicated in decreased β-cell function. However, these genetic variations have a modest effect and their combination only explains less than 10% of the T2DM heritability. With the advent of the next-generation sequencing technology, we will soon identify rare variants of larger effect as well as causal variants. These advances in understanding the genetics of T2DM will lead to the development of new therapeutic and preventive strategies and individualized medicine.
Type 2 diabetes mellitus (T2DM) is a multi-factorial disease caused by complex interplay between genetic predisposition and environmental factors [1]. Environmental factors such as increased calorie intake, physical inactivity or obesity certainly contribute to the recent diabetes epidemic. However, genetic factors are also key determinants of the individual susceptibility to T2DM. The importance of genetic risk factors for T2DM is supported by two major findings. First, there are ethnic differences in the prevalence of T2DM. Asians or Pima Indians residing in Western countries have at least a two-fold increased risk of T2DM compared to European natives [2]. Second, there is strong family history of T2DM. The offspring of T2DM parents have 40% chance of having T2DM, which is a 6-fold increased risk compared to a population risk of 7% [2].
There has been a tremendous effort to reveal the genetic predisposition of T2DM over the past 30 years or so. The three approaches adopted for identifying genetic risk factors include: 1) focusing on linkage peaks from family studies, 2) targeting candidate genes on the biological basis, and 3) genome-wide association analysis. Since the recent advent of genome-wide association studies (GWAS), there has been remarkable progress in our understanding of the genetic basis of T2DM. More than 40 genetic variations that modify the risk of T2DM development have been identified. In this article, I will review the current approaches and progress in understanding the genetics of T2DM.
Family-based linkage analysis relies on genetic markers in a family pedigree to identify the chromosomal regions showing linkage with T2DM. This approach is most useful when the disease under investigation follows a monogenic form of inheritance. Causative genetic variations of several monogenic forms of diabetes, including maturity-onset diabetes of the young, neonatal diabetes and maternally inherited diabetes and deafness were successfully identified using this approach [3-5]. However, this approach failed to identify causative genes for the common form of T2DM. There have been several reports suggesting linkage peaks near CAPN10 and ACRP30 [6,7]. Unfortunately, these results were not consistent across the study population and no high-risk variation was found to be associated with T2DM near these regions.
The candidate gene approach refers to case-control association studies focusing on specific candidate gene or a selected genetic region, chosen based on known biological function. More than hundreds of candidate genes have been investigated by this approach. However, only a few genes such as PPARG and KCNJ11 have been shown to be associated with T2DM [8,9].
The PPARG gene encodes the peroxisome proliferator-activated receptor γ, which plays a fundamental role in adipogenesis and insulin sensitivity by regulating transcriptional activity of various genes. A variation with proline at the 12th amino acid (P12A) was confirmed to be associated with a modest (odds ratio [OR] of 1.25), but significant increase in T2DM risk [8].
The KCNJ11 gene, located on the short arm of chromosome 11, encodes the pore-forming subunit of the ATP-sensitive potassium channel Kir6.2 of the pancreatic β-cells. Gain-of-function mutations of KCNJ11 open the potassium channel and inhibit the depolarization of β-cells, leading to a defect in insulin secretion [10]. Studies in various populations have consistently reported that substitution of lysine for glutamic acid at the 23rd amino acid (E23K) is associated with an increased risk of T2DM [11,12]. In recent reports of large-scale association studies and meta-analyses, the E23K variation was found to increase the risk of T2DM with an OR of 1.15 [9].
The most significant finding derived from the candidate gene approach is the strong association of TCF7L2 gene [13]. This gene was first discovered to increase the risk of T2DM through an effort to pinpoint the previously reported linkage peak [14]. The association between rs7903146T, an intronic variation, with T2DM was replicated in almost all ethnic groups and revealed to have the strongest effect in Europeans (OR, 1.46) [15-17]. A global meta-analysis showed that this variation had an OR of 1.45 and P-value of 5.4×10-140 in a comparison of more than 46,000 cases and controls [18]. However, this allele had a significantly lower allele frequency in Asians and the resultant association was much weaker [19]. The functional role of TCF7L2 in the pathogenesis of T2DM is currently under thorough investigation. It is a crucial component of Wnt signaling and is implicated in β-cell proliferation and insulin secretion [20,21]. Furthermore, a recent study reported that TCF7L2 is important in maintaining the incretin effect [22].
In the year 2007, there was a major breakthrough in identifying genetic risk factors of T2DM through the completion of GWAS. In GWAS, hundreds of thousands of single-nucleotide polymorphisms (SNPs) are tested for association with a disease, such as T2DM, in hundreds to thousands of individuals [23]. The three major advances that enabled this approach were 1) the improved knowledge of human genetic variations through the International HapMap project, 2) the technical advances in microarray genotyping methods, and 3) the progress in developing biostatistic methods to handle the large amount of data being produced. From the first six GWAS results reported in 2007, more than ten new genetic loci were reported to modify the risk of T2DM with genome-wide significance in Europeans [24-29]. These included variations in or near CDKAL1, CDKN2A/2B, SLC30A8, IGF2BP2, HHEX, and FTO. The variation in TCF7L2 was confirmed to have the strongest association in Europeans. In addition, the known association between variations in KCNJ11 and PPARG with T2DM were also replicated.
Based on the first GWAS reported by Sladek et al. [24], variations in SLC30A8 and near HHEX were found to be significantly associated with T2DM. Genetic variation of SLC30A8 is located in 8q24 and encodes an islet-specific zinc membrane transporter (ZnT8), which takes part in insulin synthesis and secretion [24]. Interestingly, the variation in SLC30A8, rs-13266634, results in a non-synonymous mutation of the protein [24]. One locus at 10q23-25 with large linkage disequilibrium block encompassing HHEX, IDE, and KIF11 were also significantly associated with T2DM. We have reported that variations in IDE, which encodes insulin degrading enzyme, are associated with the risk of T2DM in Koreans, as well as in a meta-analysis [30]. From the DGI study and FUSION study, variations in IGF2BP2 and CDKN2A/2B were found to be significantly associated with T2DM [25,27]. The IGF2BP2 gene located at 3q28, encodes insulin-like growth factor 2 mRNA-binding protein, which is thought to be involved in insulin signaling. A genetic variation located at 9p21 between CDKN2A and CDKN2B genes was also associated with T2DM. CDKN2A encodes p16INK4a and its overexpression results in decreased β-cell mass in senescent mice [31]. Another variation near this locus was independently associated with the risk of coronary heart disease due to elevated low density lipoprotein cholesterol concentration [32,33]. A genetic variation located in 6p22.3 of the CDKAL1 gene was first discovered to be significantly associated with T2DM through the deCODE study [28]. CDKAL1 encodes CDK5 regulatory subunit-associated protein 1-like 1 [28], which is thought to inhibit cyclin-dependent kinase 5 (CDK5) activity by binding to the CDK5 activator p35 [34]. In a recent report, disruption of CDKAL1 in mouse β-cells resulted in impaired first-phase insulin secretion [35]. Subjects who had risk variants of CDKAL1 had decreased insulin secretion capacity.
After the initial GWAS results, meta-analyses with large-scale replication analyses were performed [36,37]. The DIAGRAM consortium, which includes more than 50,000 cases and controls, was able to identify six additional genetic loci associated with T2DM [36]. These included variations in or near JAZF1, CDC123-CDMK1D, TSPAN8-LGR5, THADA, ADAMTS9, NOTCH2 genes. It should be noted that GWAS carried out in Asians revealed new T2DM genetic loci that were not previously reported in Europeans [38-40]. These were in or near KCNQ1, UBE2E2 and C2CD4A/4B. The KCNQ1 gene encodes a subunit of a voltage-gated potassium channel that is expressed in β-cells. The variation in KCNQ1 is thought to modulate the risk of T2DM by inducing β-cell dysfunction [38,39]. The UBE2E2 gene encodes the ubiquitin-conjugating enzyme E2E, which plays an important role in insulin synthesis and secretion under conditions where endoplasmic reticulum stress is increased in β-cells [40]. In an effort to find common genetic variations affecting fasting plasma glucose, variation in MTNR1B was found to be significantly associated with elevated fasting glucose and risk of T2DM [41-43]. MTNR1B encodes the melatonin receptor 2 (MT2) and it is expressed in human pancreatic β-cells as well as in the brain [42]. Increased expression of MT2 in β-cells is thought to suppress glucose-stimulated insulin secretion [44]. As of the end of 2010, the results from the DIAGRAM+ study, with a sample size of 141,000, included an additional 12 genetic variations, adding up to a total of more than 40 confirmed T2DM genetic risk loci [37].
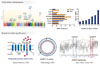
During the past ten years, my colleagues and I have focused on the candidate gene approach to find susceptibility genes for T2DM in Koreans. From these efforts, I have reported that variations in many genes, e.g., KCNJ11, PPARG, NRF1 and IDE are associated with T2DM (Table 1) [30,45-47]. We also have investigated the association of candidate genetic variations with quantitative metabolic traits such as fasting glucose, blood pressure, body mass index and dyslipidemia (Table 1) [45,48,49]. In addition, we have shown that mtDNA 16189T>C variation is associated with an increased risk of T2DM in Asians [50,51]. Among the mtDNA haplogroups, we showed that mitochondrial haplogroup N9a was significantly associated with resistance against T2DM, while D5 was significantly associated with a risk for T2DM [52]. Association of mito-chondrial DNA variation with T2DM was not observed in a European population [53,54]. For those genes identified by GWAS in Caucasians, we also confirmed the significant association between these gene variants with T2DM in Koreans (Table 2). Interestingly, the frequencies of risk alleles in Koreans were quite different from those of Caucasians, although the odds ratios of each gene variant were similar to those reported in Caucasians. These results suggest that there are important, but different contributions from genetic variants for T2DM between Caucasians and Asians (or Koreans). Fig. 1 summarizes the susceptibility genes of T2DM in the Korean population.
It is evident that the success of the GWAS approach has revolutionized our understanding of the genetic risk factors of T2DM and has provided us with better insight into the pathogenesis of T2DM. Most of the genes identified from the initial T2DM GWAS were implicated in a defect in β-cell function, rather than increased insulin resistance. It should be noted that most genetic variations only have a modest effect and their combination only explains less than 10% of the T2DM heritability [23]. The underlying hypothesis based on the current GWAS and large-scale association studies is that common genetic variations are the cause of common complex disorders. Most of the common variations with a higher than 5% frequency reside in introns or inter-genic areas and they have a relatively small effect size.
The current and future strategies to identify T2DM risk loci include performing even larger GWAS in different ethnicities. This will lead to an increase in the number of common variations that are associated with T2DM, although the size of their effects is small. Finding rare variations with larger effect size is a rational strategy to discover additional T2DM genes. Deep sequencing around the GWAS signal might yield multiple rare variations that have functional consequences. With the advent of next generation sequencing technologies, it is now possible to perform whole exome sequencing. This will allow us to identify rare functional variation with large impact on a whole exome scale. Family studies will be used more frequently as rare variants will be enriched in the relatives of the index case and the affected status would be segregated by the causative genetic variation.
Although the progress in understanding the genetics of T2DM has already been immense, it seems that this is just the beginning of a new era. Further improvements in our understanding of T2DM genetics will eventually lead us to the development of new therapeutic and preventive methods as well as the basis for individualized medicine.
Figures and Tables
Fig. 1
Susceptibility genes of type 2 diabetes mellitus in Koreans (modified from Textbook of endocrinology and metabolism. Korean Endocrine Society. 2nd ed. Seoul: Koonja Publishing; 2011. Section 9, Genetics of diabetes mellitus; p641-4. Used with permission) [63].
Notes
The Sulwon Award for Scientific Achievement is the Korean Diabetes Association's highest scientific award and honors an individual who has excellently contributed to the progress in the field of diabetes and metabolism. Sulwon award is named after an emeritus professor Eung Jin Kim, who founded Korean Diabetes Association.
Prof. Kyong Soo Park received the first Sulwon Award at 35th Autumn Congress of Korean Diabetes Association, Nov 19-21, 2009 at Jeju, Korea.
References
1. Stumvoll M, Goldstein BJ, van Haeften TW. Type 2 diabetes: principles of pathogenesis and therapy. Lancet. 2005. 365:1333–1346.
2. Doria A, Patti ME, Kahn CR. The emerging genetic architecture of type 2 diabetes. Cell Metab. 2008. 8:186–200.
3. Froguel P, Vaxillaire M, Sun F, Velho G, Zouali H, Butel MO, Lesage S, Vionnet N, Clement K, Fougerousse F, Tanizawa Y, Weissenbach J, Beckmann JS, Lathrop GM, Passa PH, Permutt MA, Cohen D. Close linkage of glucokinase locus on chromosome 7p to early-onset non-insulin-dependent diabetes mellitus. Nature. 1992. 356:162–164.
4. Vaxillaire M, Froguel P. Monogenic diabetes in the young, pharmacogenetics and relevance to multifactorial forms of type 2 diabetes. Endocr Rev. 2008. 29:254–264.
5. Greeley SA, Tucker SE, Worrell HI, Skowron KB, Bell GI, Philipson LH. Update in neonatal diabetes. Curr Opin Endocrinol Diabetes Obes. 2010. 17:13–19.
6. Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, Hara M, Hinokio Y, Lindner TH, Mashima H, Schwarz PE, del Bosque-Plata L, Horikawa Y, Oda Y, Yoshiuchi I, Colilla S, Polonsky KS, Wei S, Concannon P, Iwasaki N, Schulze J, Baier LJ, Bogardus C, Groop L, Boerwinkle E, Hanis CL, Bell GI. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet. 2000. 26:163–175.
7. Vionnet N, Hani EH, Dupont S, Gallina S, Francke S, Dotte S, De Matos F, Durand E, Lepretre F, Lecoeur C, Gallina P, Zekiri L, Dina C, Froguel P. Genomewide search for type 2 diabetes-susceptibility genes in French whites: evidence for a novel susceptibility locus for early-onset diabetes on chromosome 3q27-qter and independent replication of a type 2-diabetes locus on chromosome 1q21-q24. Am J Hum Genet. 2000. 67:1470–1480.
8. Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, Tuomi T, Gaudet D, Hudson TJ, Daly M, Groop L, Lander ES. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000. 26:76–80.
9. Gloyn AL, Weedon MN, Owen KR, Turner MJ, Knight BA, Hitman G, Walker M, Levy JC, Sampson M, Halford S, McCarthy MI, Hattersley AT, Frayling TM. Large-scale association studies of variants in genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes. 2003. 52:568–572.
10. Gloyn AL, Pearson ER, Antcliff JF, Proks P, Bruining GJ, Slingerland AS, Howard N, Srinivasan S, Silva JM, Molnes J, Edghill EL, Frayling TM, Temple IK, Mackay D, Shield JP, Sumnik Z, van Rhijn A, Wales JK, Clark P, Gorman S, Aisenberg J, Ellard S, Njolstad PR, Ashcroft FM, Hattersley AT. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N Engl J Med. 2004. 350:1838–1849.
11. Florez JC, Burtt N, de Bakker PI, Almgren P, Tuomi T, Holmkvist J, Gaudet D, Hudson TJ, Schaffner SF, Daly MJ, Hirschhorn JN, Groop L, Altshuler D. Haplotype structure and genotype-phenotype correlations of the sulfonylurea receptor and the islet ATP-sensitive potassium channel gene region. Diabetes. 2004. 53:1360–1368.
12. Nielsen EM, Hansen L, Carstensen B, Echwald SM, Drivsholm T, Glumer C, Thorsteinsson B, Borch-Johnsen K, Hansen T, Pedersen O. The E23K variant of Kir6.2 associates with impaired post-OGTT serum insulin response and increased risk of type 2 diabetes. Diabetes. 2003. 52:573–577.
13. Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, Styrkarsdottir U, Magnusson KP, Walters GB, Palsdottir E, Jonsdottir T, Gudmundsdottir T, Gylfason A, Saemundsdottir J, Wilensky RL, Reilly MP, Rader DJ, Bagger Y, Christiansen C, Gudnason V, Sigurdsson G, Thorsteinsdottir U, Gulcher JR, Kong A, Stefansson K. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006. 38:320–323.
14. Reynisdottir I, Thorleifsson G, Benediktsson R, Sigurdsson G, Emilsson V, Einarsdottir AS, Hjorleifsdottir EE, Orlygsdottir GT, Bjornsdottir GT, Saemundsdottir J, Halldorsson S, Hrafnkelsdottir S, Sigurjonsdottir SB, Steinsdottir S, Martin M, Kochan JP, Rhees BK, Grant SF, Frigge ML, Kong A, Gudnason V, Stefansson K, Gulcher JR. Localization of a susceptibility gene for type 2 diabetes to chromosome 5q34-q35.2. Am J Hum Genet. 2003. 73:323–335.
15. Florez JC, Jablonski KA, Bayley N, Pollin TI, de Bakker PI, Shuldiner AR, Knowler WC, Nathan DM, Altshuler D. Diabetes Prevention Program Research Group. TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. N Engl J Med. 2006. 355:241–250.
16. Saxena R, Gianniny L, Burtt NP, Lyssenko V, Giuducci C, Sjogren M, Florez JC, Almgren P, Isomaa B, Orho-Melander M, Lindblad U, Daly MJ, Tuomi T, Hirschhorn JN, Ardlie KG, Groop LC, Altshuler D. Common single nucleotide polymorphisms in TCF7L2 are reproducibly associated with type 2 diabetes and reduce the insulin response to glucose in nondiabetic individuals. Diabetes. 2006. 55:2890–2895.
17. Chang YC, Chang TJ, Jiang YD, Kuo SS, Lee KC, Chiu KC, Chuang LM. Association study of the genetic polymorphisms of the transcription factor 7-like 2 (TCF7L2) gene and type 2 diabetes in the Chinese population. Diabetes. 2007. 56:2631–2637.
18. Cauchi S, El Achhab Y, Choquet H, Dina C, Krempler F, Weitgasser R, Nejjari C, Patsch W, Chikri M, Meyre D, Froguel P. TCF7L2 is reproducibly associated with type 2 diabetes in various ethnic groups: a global meta-analysis. J Mol Med. 2007. 85:777–782.
19. Ng MC, Park KS, Oh B, Tam CH, Cho YM, Shin HD, Lam VK, Ma RC, So WY, Cho YS, Kim HL, Lee HK, Chan JC, Cho NH. Implication of genetic variants near TCF7L2, SLC30A8, HHEX, CDKAL1, CDKN2A/B, IGF2BP2, and FTO in type 2 diabetes and obesity in 6,719 Asians. Diabetes. 2008. 57:2226–2233.
20. Lyssenko V, Lupi R, Marchetti P, Del Guerra S, Orho-Melander M, Almgren P, Sjogren M, Ling C, Eriksson KF, Lethagen AL, Mancarella R, Berglund G, Tuomi T, Nilsson P, Del Prato S, Groop L. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest. 2007. 117:2155–2163.
21. Shu L, Sauter NS, Schulthess FT, Matveyenko AV, Oberholzer J, Maedler K. Transcription factor 7-like 2 regulates beta-cell survival and function in human pancreatic islets. Diabetes. 2008. 57:645–653.
22. Schafer SA, Tschritter O, Machicao F, Thamer C, Stefan N, Gallwitz B, Holst JJ, Dekker JM, 't Hart LM, Nijpels G, van Haeften TW, Haring HU, Fritsche A. Impaired glucagon-like peptide-1-induced insulin secretion in carriers of transcription factor 7-like 2 (TCF7L2) gene polymorphisms. Diabetologia. 2007. 50:2443–2450.
23. Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009. 461:747–753.
24. Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007. 445:881–885.
25. Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007. 316:1341–1345.
26. Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, Barrett JC, Shields B, Morris AP, Ellard S, Groves CJ, Harries LW, Marchini JL, Owen KR, Knight B, Cardon LR, Walker M, Hitman GA, Morris AD, Doney AS, McCarthy MI, Hattersley AT. Wellcome Trust Case Control Consortium (WTCCC). Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007. 316:1336–1341.
27. Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research. Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Bostrom K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Rastam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjogren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007. 316:1331–1336.
28. Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, Walters GB, Styrkarsdottir U, Gretarsdottir S, Emilsson V, Ghosh S, Baker A, Snorradottir S, Bjarnason H, Ng MC, Hansen T, Bagger Y, Wilensky RL, Reilly MP, Adeyemo A, Chen Y, Zhou J, Gudnason V, Chen G, Huang H, Lashley K, Doumatey A, So WY, Ma RC, Andersen G, Borch-Johnsen K, Jorgensen T, van Vliet-Ostaptchouk JV, Hofker MH, Wijmenga C, Christiansen C, Rader DJ, Rotimi C, Gurney M, Chan JC, Pedersen O, Sigurdsson G, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007. 39:770–775.
29. Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007. 447:661–678.
30. Kwak SH, Cho YM, Moon MK, Kim JH, Park BL, Cheong HS, Shin HD, Jang HC, Kim SY, Lee HK, Park KS. Association of polymorphisms in the insulin-degrading enzyme gene with type 2 diabetes in the Korean population. Diabetes Res Clin Pract. 2008. 79:284–290.
31. Krishnamurthy J, Ramsey MR, Ligon KL, Torrice C, Koh A, Bonner-Weir S, Sharpless NE. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006. 443:453–457.
32. Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G, Gudbjartsson DF, Magnusson KP, Andersen K, Levey AI, Backman VM, Matthiasdottir S, Jonsdottir T, Palsson S, Einarsdottir H, Gunnarsdottir S, Gylfason A, Vaccarino V, Hooper WC, Reilly MP, Granger CB, Austin H, Rader DJ, Shah SH, Quyyumi AA, Gulcher JR, Thorgeirsson G, Thorsteinsdottir U, Kong A, Stefansson K. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007. 316:1491–1493.
33. McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007. 316:1488–1491.
34. Wei FY, Nagashima K, Ohshima T, Saheki Y, Lu YF, Matsushita M, Yamada Y, Mikoshiba K, Seino Y, Matsui H, Tomizawa K. Cdk5-dependent regulation of glucose-stimulated insulin secretion. Nat Med. 2005. 11:1104–1108.
35. Ohara-Imaizumi M, Yoshida M, Aoyagi K, Saito T, Okamura T, Takenaka H, Akimoto Y, Nakamichi Y, Takanashi-Yanobu R, Nishiwaki C, Kawakami H, Kato N, Hisanaga S, Kakei M, Nagamatsu S. Deletion of CDKAL1 affects mitochondrial ATP generation and first-phase insulin exocytosis. PLoS One. 2010. 5:e15553.
36. Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, de Bakker PI, Abecasis GR, Almgren P, Andersen G, Ardlie K, Bostrom KB, Bergman RN, Bonnycastle LL, Borch-Johnsen K, Burtt NP, Chen H, Chines PS, Daly MJ, Deodhar P, Ding CJ, Doney AS, Duren WL, Elliott KS, Erdos MR, Frayling TM, Freathy RM, Gianniny L, Grallert H, Grarup N, Groves CJ, Guiducci C, Hansen T, Herder C, Hitman GA, Hughes TE, Isomaa B, Jackson AU, Jorgensen T, Kong A, Kubalanza K, Kuruvilla FG, Kuusisto J, Langenberg C, Lango H, Lauritzen T, Li Y, Lindgren CM, Lyssenko V, Marvelle AF, Meisinger C, Midthjell K, Mohlke KL, Morken MA, Morris AD, Narisu N, Nilsson P, Owen KR, Palmer CN, Payne F, Perry JR, Pettersen E, Platou C, Prokopenko I, Qi L, Qin L, Rayner NW, Rees M, Roix JJ, Sandbaek A, Shields B, Sjogren M, Steinthorsdottir V, Stringham HM, Swift AJ, Thorleifsson G, Thorsteinsdottir U, Timpson NJ, Tuomi T, Tuomilehto J, Walker M, Watanabe RM, Weedon MN, Willer CJ. Wellcome Trust Case Control Consortium. Illig T, Hveem K, Hu FB, Laakso M, Stefansson K, Pedersen O, Wareham NJ, Barroso I, Hattersley AT, Collins FS, Groop L, McCarthy MI, Boehnke M, Altshuler D. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008. 40:638–645.
37. Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, Lindgren CM, Magi R, Morris AP, Randall J, Johnson T, Elliott P, Rybin D, Thorleifsson G, Steinthorsdottir V, Henneman P, Grallert H, Dehghan A, Hottenga JJ, Franklin CS, Navarro P, Song K, Goel A, Perry JR, Egan JM, Lajunen T, Grarup N, Sparso T, Doney A, Voight BF, Stringham HM, Li M, Kanoni S, Shrader P, Cavalcanti-Proenca C, Kumari M, Qi L, Timpson NJ, Gieger C, Zabena C, Rocheleau G, Ingelsson E, An P, O'Connell J, Luan J, Elliott A, McCarroll SA, Payne F, Roccasecca RM, Pattou F, Sethupathy P, Ardlie K, Ariyurek Y, Balkau B, Barter P, Beilby JP, Ben-Shlomo Y, Benediktsson R, Bennett AJ, Bergmann S, Bochud M, Boerwinkle E, Bonnefond A, Bonnycastle LL, Borch-Johnsen K, Bottcher Y, Brunner E, Bumpstead SJ, Charpentier G, Chen YD, Chines P, Clarke R, Coin LJ, Cooper MN, Cornelis M, Crawford G, Crisponi L, Day IN, de Geus EJ, Delplanque J, Dina C, Erdos MR, Fedson AC, Fischer-Rosinsky A, Forouhi NG, Fox CS, Frants R, Franzosi MG, Galan P, Goodarzi MO, Graessler J, Groves CJ, Grundy S, Gwilliam R, Gyllensten U, Hadjadj S, Hallmans G, Hammond N, Han X, Hartikainen AL, Hassanali N, Hayward C, Heath SC, Hercberg S, Herder C, Hicks AA, Hillman DR, Hingorani AD, Hofman A, Hui J, Hung J, Isomaa B, Johnson PR, Jorgensen T, Jula A, Kaakinen M, Kaprio J, Kesaniemi YA, Kivimaki M, Knight B, Koskinen S, Kovacs P, Kyvik KO, Lathrop GM, Lawlor DA, Le Bacquer O, Lecoeur C, Li Y, Lyssenko V, Mahley R, Mangino M, Manning AK, Martinez-Larrad MT, McAteer JB, McCulloch LJ, McPherson R, Meisinger C, Melzer D, Meyre D, Mitchell BD, Morken MA, Mukherjee S, Naitza S, Narisu N, Neville MJ, Oostra BA, Orru M, Pakyz R, Palmer CN, Paolisso G, Pattaro C, Pearson D, Peden JF, Pedersen NL, Perola M, Pfeiffer AF, Pichler I, Polasek O, Posthuma D, Potter SC, Pouta A, Province MA, Psaty BM, Rathmann W, Rayner NW, Rice K, Ripatti S, Rivadeneira F, Roden M, Rolandsson O, Sandbaek A, Sandhu M, Sanna S, Sayer AA, Scheet P, Scott LJ, Seedorf U, Sharp SJ, Shields B, Sigurethsson G, Sijbrands EJ, Silveira A, Simpson L, Singleton A, Smith NL, Sovio U, Swift A, Syddall H, Syvanen AC, Tanaka T, Thorand B, Tichet J, Tonjes A, Tuomi T, Uitterlinden AG, van Dijk KW, van Hoek M, Varma D, Visvikis-Siest S, Vitart V, Vogelzangs N, Waeber G, Wagner PJ, Walley A, Walters GB, Ward KL, Watkins H, Weedon MN, Wild SH, Willemsen G, Witteman JC, Yarnell JW, Zeggini E, Zelenika D, Zethelius B, Zhai G, Zhao JH, Zillikens MC. DIAGRAM Consortium. GIANT Consortium. Global BPgen Consortium. Borecki IB, Loos RJ, Meneton P, Magnusson PK, Nathan DM, Williams GH, Hattersley AT, Silander K, Salomaa V, Smith GD, Bornstein SR, Schwarz P, Spranger J, Karpe F, Shuldiner AR, Cooper C, Dedoussis GV, Serrano-Rios M, Morris AD, Lind L, Palmer LJ, Hu FB, Franks PW, Ebrahim S, Marmot M, Kao WH, Pankow JS, Sampson MJ, Kuusisto J, Laakso M, Hansen T, Pedersen O, Pramstaller PP, Wichmann HE, Illig T, Rudan I, Wright AF, Stumvoll M, Campbell H, Wilson JF. Anders Hamsten on behalf of Procardis Consortium. MAGIC investigators. Bergman RN, Buchanan TA, Collins FS, Mohlke KL, Tuomilehto J, Valle TT, Altshuler D, Rotter JI, Siscovick DS, Penninx BW, Boomsma DI, Deloukas P, Spector TD, Frayling TM, Ferrucci L, Kong A, Thorsteinsdottir U, Stefansson K, van Duijn CM, Aulchenko YS, Cao A, Scuteri A, Schlessinger D, Uda M, Ruokonen A, Jarvelin MR, Waterworth DM, Vollenweider P, Peltonen L, Mooser V, Abecasis GR, Wareham NJ, Sladek R, Froguel P, Watanabe RM, Meigs JB, Groop L, Boehnke M, McCarthy MI, Florez JC, Barroso I. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010. 42:105–116.
38. Yasuda K, Miyake K, Horikawa Y, Hara K, Osawa H, Furuta H, Hirota Y, Mori H, Jonsson A, Sato Y, Yamagata K, Hinokio Y, Wang HY, Tanahashi T, Nakamura N, Oka Y, Iwasaki N, Iwamoto Y, Yamada Y, Seino Y, Maegawa H, Kashiwagi A, Takeda J, Maeda E, Shin HD, Cho YM, Park KS, Lee HK, Ng MC, Ma RC, So WY, Chan JC, Lyssenko V, Tuomi T, Nilsson P, Groop L, Kamatani N, Sekine A, Nakamura Y, Yamamoto K, Yoshida T, Tokunaga K, Itakura M, Makino H, Nanjo K, Kadowaki T, Kasuga M. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet. 2008. 40:1092–1097.
39. Unoki H, Takahashi A, Kawaguchi T, Hara K, Horikoshi M, Andersen G, Ng DP, Holmkvist J, Borch-Johnsen K, Jorgensen T, Sandbaek A, Lauritzen T, Hansen T, Nurbaya S, Tsunoda T, Kubo M, Babazono T, Hirose H, Hayashi M, Iwamoto Y, Kashiwagi A, Kaku K, Kawamori R, Tai ES, Pedersen O, Kamatani N, Kadowaki T, Kikkawa R, Nakamura Y, Maeda S. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet. 2008. 40:1098–1102.
40. Yamauchi T, Hara K, Maeda S, Yasuda K, Takahashi A, Horikoshi M, Nakamura M, Fujita H, Grarup N, Cauchi S, Ng DP, Ma RC, Tsunoda T, Kubo M, Watada H, Maegawa H, Okada-Iwabu M, Iwabu M, Shojima N, Shin HD, Andersen G, Witte DR, Jorgensen T, Lauritzen T, Sandbaek A, Hansen T, Ohshige T, Omori S, Saito I, Kaku K, Hirose H, So WY, Beury D, Chan JC, Park KS, Tai ES, Ito C, Tanaka Y, Kashiwagi A, Kawamori R, Kasuga M, Froguel P, Pedersen O, Kamatani N, Nakamura Y, Kadowaki T. A genome-wide association study in the Japanese population identifies susceptibility loci for type 2 diabetes at UBE2E2 and C2CD4A-C2CD4B. Nat Genet. 2010. 42:864–868.
41. Bouatia-Naji N, Bonnefond A, Cavalcanti-Proenca C, Sparso T, Holmkvist J, Marchand M, Delplanque J, Lobbens S, Rocheleau G, Durand E, De Graeve F, Chevre JC, Borch-Johnsen K, Hartikainen AL, Ruokonen A, Tichet J, Marre M, Weill J, Heude B, Tauber M, Lemaire K, Schuit F, Elliott P, Jorgensen T, Charpentier G, Hadjadj S, Cauchi S, Vaxillaire M, Sladek R, Visvikis-Siest S, Balkau B, Levy-Marchal C, Pattou F, Meyre D, Blakemore AI, Jarvelin MR, Walley AJ, Hansen T, Dina C, Pedersen O, Froguel P. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet. 2009. 41:89–94.
42. Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, Thorleifsson G, Loos RJ, Manning AK, Jackson AU, Aulchenko Y, Potter SC, Erdos MR, Sanna S, Hottenga JJ, Wheeler E, Kaakinen M, Lyssenko V, Chen WM, Ahmadi K, Beckmann JS, Bergman RN, Bochud M, Bonnycastle LL, Buchanan TA, Cao A, Cervino A, Coin L, Collins FS, Crisponi L, de Geus EJ, Dehghan A, Deloukas P, Doney AS, Elliott P, Freimer N, Gateva V, Herder C, Hofman A, Hughes TE, Hunt S, Illig T, Inouye M, Isomaa B, Johnson T, Kong A, Krestyaninova M, Kuusisto J, Laakso M, Lim N, Lindblad U, Lindgren CM, McCann OT, Mohlke KL, Morris AD, Naitza S, Orru M, Palmer CN, Pouta A, Randall J, Rathmann W, Saramies J, Scheet P, Scott LJ, Scuteri A, Sharp S, Sijbrands E, Smit JH, Song K, Steinthorsdottir V, Stringham HM, Tuomi T, Tuomilehto J, Uitterlinden AG, Voight BF, Waterworth D, Wichmann HE, Willemsen G, Witteman JC, Yuan X, Zhao JH, Zeggini E, Schlessinger D, Sandhu M, Boomsma DI, Uda M, Spector TD, Penninx BW, Altshuler D, Vollenweider P, Jarvelin MR, Lakatta E, Waeber G, Fox CS, Peltonen L, Groop LC, Mooser V, Cupples LA, Thorsteinsdottir U, Boehnke M, Barroso I, Van Duijn C, Dupuis J, Watanabe RM, Stefansson K, McCarthy MI, Wareham NJ, Meigs JB, Abecasis GR. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009. 41:77–81.
43. Lyssenko V, Nagorny CL, Erdos MR, Wierup N, Jonsson A, Spegel P, Bugliani M, Saxena R, Fex M, Pulizzi N, Isomaa B, Tuomi T, Nilsson P, Kuusisto J, Tuomilehto J, Boehnke M, Altshuler D, Sundler F, Eriksson JG, Jackson AU, Laakso M, Marchetti P, Watanabe RM, Mulder H, Groop L. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet. 2009. 41:82–88.
44. Mulder H, Nagorny CL, Lyssenko V, Groop L. Melatonin receptors in pancreatic islets: good morning to a novel type 2 diabetes gene. Diabetologia. 2009. 52:1240–1249.
45. Koo BK, Cho YM, Park BL, Cheong HS, Shin HD, Jang HC, Kim SY, Lee HK, Park KS. Polymorphisms of KCNJ11 (Kir6.2 gene) are associated with type 2 diabetes and hypertension in the Korean population. Diabet Med. 2007. 24:178–186.
46. Moon MK, Cho YM, Jung HS, Park YJ, Yoon KH, Sung YA, Park BL, Lee HK, Park KS, Shin HD. Genetic polymorphisms in peroxisome proliferator-activated receptor gamma are associated with type 2 diabetes mellitus and obesity in the Korean population. Diabet Med. 2005. 22:1161–1166.
47. Cho YM, Shin HD, Park BL, Kim JH, Park KS, Kim SY, Lee HK. Association between polymorphisms in the nuclear respiratory factor 1 gene and type 2 diabetes mellitus in the Korean population. Diabetologia. 2005. 48:2033–2038.
48. Choi HJ, Cho YM, Moon MK, Choi HH, Shin HD, Jang HC, Kim SY, Lee HK, Park KS. Polymorphisms in the ghrelin gene are associated with serum high-density lipoprotein cholesterol level and not with type 2 diabetes mellitus in Koreans. J Clin Endocrinol Metab. 2006. 91:4657–4663.
49. Shin HD, Park BL, Kim LH, Jung HS, Cho YM, Moon MK, Park YJ, Lee HK, Park KS. Genetic polymorphisms in peroxisome proliferator-activated receptor delta associated with obesity. Diabetes. 2004. 53:847–851.
50. Kim JH, Park KS, Cho YM, Kang BS, Kim SK, Jeon HJ, Kim SY, Lee HK. The prevalence of the mitochondrial DNA 16189 variant in non-diabetic Korean adults and its association with higher fasting glucose and body mass index. Diabet Med. 2002. 19:681–684.
51. Park KS, Chan JC, Chuang LM, Suzuki S, Araki E, Nanjo K, Ji L, Ng M, Nishi M, Furuta H, Shirotani T, Ahn BY, Chung SS, Min HK, Lee SW, Kim JH, Cho YM, Lee HK. Study Group of Molecular Diabetology in Asia. A mitochondrial DNA variant at position 16189 is associated with type 2 diabetes mellitus in Asians. Diabetologia. 2008. 51:602–608.
52. Fuku N, Park KS, Yamada Y, Nishigaki Y, Cho YM, Matsuo H, Segawa T, Watanabe S, Kato K, Yokoi K, Nozawa Y, Lee HK, Tanaka M. Mitochondrial haplogroup N9a confers resistance against type 2 diabetes in Asians. Am J Hum Genet. 2007. 80:407–415.
53. Chinnery PF, Elliott HR, Patel S, Lambert C, Keers SM, Durham SE, McCarthy MI, Hitman GA, Hattersley AT, Walker M. Role of the mitochondrial DNA 16184-16193 poly-C tract in type 2 diabetes. Lancet. 2005. 366:1650–1651.
54. Saxena R, de Bakker PI, Singer K, Mootha V, Burtt N, Hirschhorn JN, Gaudet D, Isomaa B, Daly MJ, Groop L, Ardlie KG, Altshuler D. Comprehensive association testing of common mitochondrial DNA variation in metabolic disease. Am J Hum Genet. 2006. 79:54–61.
55. Kim JH, Shin HD, Park BL, Cho YM, Kim SY, Lee HK, Park KS. Peroxisome proliferator-activated receptor gamma coactivator 1 alpha promoter polymorphisms are associated with early-onset type 2 diabetes mellitus in the Korean population. Diabetologia. 2005. 48:1323–1330.
56. Park KS, Shin HD, Park BL, Cheong HS, Cho YM, Lee HK, Lee JY, Lee JK, Kim HT, Park CS, Han BG, Kimm K, Oh B. Putative association of peroxisome proliferator-activated receptor gamma co-activator 1beta (PPARGC1B) polymorphism with type 2 diabetes mellitus. Diabet Med. 2006. 23:635–642.
57. Lee HJ, Ryu HJ, Shin HD, Park BL, Kim JY, Cho YM, Park KS, Song J, Oh B. Associations between polymorphisms in the mitochondrial uncoupling proteins (UCPs) with T2DM. Clin Chim Acta. 2008. 398:27–33.
58. Shin HD, Park KS, Park BL, Cheong HS, Cho YM, Lee HK, Lee JY, Lee JK, Kim HT, Han BG, Kim JW, Koh I, Kim YJ, Oh B, Kimm K, Park C. Common promoter polymorphism in monocyte differentiation antigen CD14 is associated with serum triglyceride levels and body mass index in non-diabetic individuals. Diabet Med. 2006. 23:72–76.
59. Ku YH, Koo BK, Kwak SH, Cho YM, Shin HD, Lee HK, Kim Y, Choi JW, Oh B, Park KS. Regulatory effect of common promoter polymorphisms on the expression of the 11beta-hydroxysteroid dehydrogenase type 1 gene. Horm Res. 2009. 72:25–32.
60. Park KS, Shin HD, Park BL, Cheong HS, Cho YM, Lee HK, Lee JY, Lee JK, Oh B, Kimm K. Polymorphisms in the leptin receptor (LEPR): putative association with obesity and T2DM. J Hum Genet. 2006. 51:85–91.
61. Kim JT, Kim Y, Cho YM, Koo BK, Lee EK, Shin HD, Jang HC, Choi JW, Oh B, Park KS. Polymorphisms of ADIPOR1 and ADIPOR2 are associated with phenotypes of type 2 diabetes in Koreans. Clin Endocrinol (Oxf). 2009. 70:66–74.
62. Shin HD, Park BL, Kim LH, Cheong HS, Kim JH, Cho YM, Lee HK, Park KS. Association of a polymorphism in the gene encoding phosphoenolpyruvate carboxykinase 1 with high-density lipoprotein and triglyceride levels. Diabetologia. 2005. 48:2025–2032.
63. Korean Endocrine Society. Section 9, Genetics of diabetes mellitus. Textbook of endocrinology and metabolism. 2011. 2nd ed. Seoul: Koonja Publishing;641–644.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download