Abstract
Allopurinol-induced severe cutaneous adverse reactions (SCARs) such as Stevens-Johnson syndrome (SJS), toxic epidermal necrolysis (TEN), and drug reaction with eosinophilia and systemic symptoms (DRESS) syndrome are reportedly associated with the HLA-B*58:01 genotype. Three patients who developed SCARs after allopurinol administration were subjected to HLA-B genotyping and lymphocyte activation test (LAT) to evaluate genetic risk and to detect the causative agent, respectively. All three patients given allopurinol to treat gout were diagnosed with DRESS syndrome. Symptom onset commenced 7-24 days after drug exposure; the patients took allopurinol (100–200 mg/d) for 2-30 days. HLA-B genotyping was performed using a polymerase chain reaction (PCR)-sequence-based typing (SBT) method. All patients had a single HLA-B*58:01 allele: HLA-B*13:02/*58:01 (a 63-year-old male), HLA-B*48:01/*58:01 (a 71-year-old female), and HLA-B*44:03/*58:01 (a 22-year-old male). Only the last patient yielded a positive LAT result, confirming that allopurinol was the causative agent. These findings suggest that patients with HLA-B*58:01 may develop SCARs upon allopurinol administration. Therefore, HLA-B genotyping could be helpful in preventing serious problems attributable to allopurinol treatment, although PCR-SBT HLA-B genotyping is time consuming. A simple genotyping test is required in practice. LAT may help to identify a causative agent.
Allopurinol is frequently used to treat hyperuricemia. However, allopurinol may induce severe cutaneous adverse reactions (SCARs); these life-threatening disorders include Stevens-Johnson syndrome (SJS), toxic epidermal necrolysis (TEN), acute generalized exanthematous pustulosis (AGEP), hypersensitivity syndrome (HSS), drug-induced hypersensitivity syndrome (DIHS), and drug reaction with eosinophilia and systemic symptoms (DRESS) syndrome.[1] Relationships between human leukocyte antigen (HLA) genotype and drug-induced hypersensitivity to abacavir, carbamazepine, and other drugs have been described.[23] Associations between the HLA-B*58:01 genotype and allopurinol-induced SCARs have been reported in various ethnic populations, including Chinese,[4] Japanese,[5] and Korean.[6] We describe our experience with three patients who developed allopurinol-induced SCARs; we review the clinical findings, HLA-B genotypes, and results of lymphocyte activation test (LAT).
Written informed consent was obtained from all three patients and the work was approved by the Institutional Review Board of Inje University Busan Paik Hospital, Busan, Republic of Korea. We collected clinical data, HLA-B genotyping results, and LAT outcomes for all patients.
Peripheral blood was collected from all patients, and genomic DNA was extracted from peripheral blood mononuclear cells (PBMCs) using a QIAamp® Blood Mini Kit (QIAGEN, Hilden, Germany). We determined HLA-B genotypes using polymerase chain reaction (PCR)-sequence-based typing (SBT) (SBT Engine software version 2.20; GenDx, Utrecht, the Netherlands).
To explore drug hypersensitivity, we subjected PBMCs to LAT.[7] Cells were also incubated with or without 0.5 µg/mL phytohemagglutinin A (PHA; positive and negative controls, respectively; Sigma-Aldrich, St. Louis, MO, USA). Briefly, we subjected allopurinol, carbamazepine, and phenytoin to LAT. Blood was layered onto Histopaque®-1077 (Sigma-Aldrich) and single-cell suspensions were obtained by centrifugation. These suspensions were added to 96-well culture plates and stimulated with several concentrations of allopurinol for 48 and 72 h. The allopurinol concentrations used were the same as those in a case report by Kim et al.[8] After incubation, the CD69 and CD25 expression levels on CD4+/CD8+ T cells were measured by flow cytometry.[9] The cells were harvested and stained with antihuman CD3 PE, anti-human CD4 PECy5, anti-human CD8 APC, anti-human CD69 FITC, and anti-human CD25 PECy7 (eBioscience, San Diego, CA, USA). Data acquired with the aid of a FACSCanto™ II flow cytometry (BD Biosciences, San Jose, CA, USA) were analyzed using FlowJo® software (Tree Star, Ashland, OR, USA). The data from stimulated samples were divided by those from unstimulated samples to obtain stimulation indices (SIs). An SI > 2.0 was interpreted as positive.
A 63-year-old male presented with a 14-day history of pruritic erythematous papules over his entire body, accompanied by jaundice. He had been using a topical corticosteroid and had taken an antihistamine for 10 days before visiting, but the symptoms became aggravated, with eye and oral mucosal involvement. To treat hypertension and gout, he had commenced anti-hypertensive drugs 5 months prior and allopurinol 3 weeks prior, respectively. A high fever (>38.0℃) was evident on physical examination. Laboratory tests revealed elevated levels of aspartate aminotransferase (AST; 161 U/L), alanine aminotransferase (ALT; 390 U/L), total bilirubin (10.9 mg/dL), and eosinophils (2,432/mm2). The histopathological data were compatible with erythema multiforme. Genetic analysis revealed the HLA-B*13:02/*58:01 genotype. We diagnosed DRESS syndrome caused by allopurinol and prescribed systemic corticosteroids, after which clinical improvement was evident.
A 71-year-old female presented with a 3-day history of painful erythematous scaly patches that were coalescing into large patches, progressive desquamation over her entire body, a febrile sensation, and general weakness. She had been taking anti-hypertensive medication for 5 years. One month prior, she had commenced allopurinol and colchicine to treat newly diagnosed gout. Her body temperature was >38.0℃. Laboratory tests revealed elevated levels of AST (281 U/L), ALT (118 U/L), and eosinophil (1,607/mm2). The histopathological data were compatible with erythema multiforme. Genetic analysis revealed the HLA-B*48:01/*58:01 genotype. We diagnosed DRESS syndrome attributable to allopurinol. She was also diagnosed with end-stage renal disease (ESRD) during work-up of the skin lesions. After 3 months of treatment with systemic and topical corticosteroids, clinical improvement was evident.
A 22-year-old male presented with a 10-day history of painful erythematous scaly patches coalescing into larger patches, progressive desquamation over the entire body, marked facial swelling, and a febrile sensation (Fig. 1). He had a history of diabetes mellitus, tuberculosis, and gout. He had taken anti-hypertensive drugs for 5 years, and had commenced anti-tuberculosis medication 4 months prior, and allopurinol 1 month prior (to treat gout). High fever (>38.0℃) and submandibular lymph node enlargement were evident on physical examination. Laboratory tests revealed elevated levels of AST (86 U/L), ALT (101 U/L), and eosinophils (2,239/mm2). Histopathological examination revealed acanthosis with severe lymphocytic exocytosis and spongiosis of the epidermis, and dense perivascular and perifollicular lymphocytic infiltration (Fig. 1). Genetic analysis revealed the HLA-B*44:03/*58:01 genotype. LAT was positive for allopurinol (Fig. 2). He was diagnosed with DRESS syndrome attributable to allopurinol and treated with cyclosporine, antipyretics, and topical corticosteroids. After 3 weeks of treatment, the clinical symptoms and signs subsided.
We diagnosed three patients with allopurinol-induced SCARs based on clinical features, pathological findings, and laboratory data; all had DRESS syndrome. All patients were taking allopurinol to treat gout and recovered from their allopurinol-induced SCARs after intensive care. All had HLA-B*58:01 alleles and LAT confirmed that allopurinol was the causative agent in one patient (Fig. 2). The patients had taken allopurinol at 100–200 mg/d for 2–30 days; the time to symptom onset after the initial dose was 7–24 days (Table 1). Allopurinol-induced hypersensitivity often co-exists with an HLA-B*58:01 allele and renal impairment.[10] Patient 2 had both an HLA-B*58:01 allele and ESRD, triggering DRESS syndrome. The other patients also had concomitant diseases: hypertension, diabetes mellitus, and tuberculosis.
Genetic markers apart from HLA-B*58:01, including HLA-C*03:02 and A*33:02 (risk factors) and A*02:01 (a preventative marker), are reportedly associated with allopurinol-induced SCARs in Korean patients.[6] We performed HLA-B genotyping only; we do not know whether the patients had other HLA risk alleles. Jutkowitz et al. found that HLA-B*58:01 testing before allopurinol prescription was cost-effective for Asians living in the USA (such as those of Korean, Thai, or Han Chinese descent),[11] and also when the population frequency of HLA-B*58:01 was >1.6%. The HLA-B*58:01 allele frequency in Koreans is 6.77%,[12] rendering this allele relatively common; it may be cost-effective to evaluate HLA-B*58:01 status before prescribing allopurinol.
LAT serves to detect drug sensitization at the cellular level in patients with non-immediate T cell-mediated drug hypersensitivity reactions; the sensitivity and specificity of LAT are 16.7% and 97.8%, respectively,[7] suggesting that LAT can be used to confirm a causative agent. Only 1 of the 3 patients was positive on LAT because of the low sensitivity of the test; however, the result was meaningful in that the causative agent was identified. Other diagnostic tests for non-immediate drug hypersensitivity reactions include the in vitro lymphocyte transformation test (LTT) and enzyme-linked immunospot (ELISPOT) assays; and in vivo delayed-reading intradermal tests, patch tests, and provocation tests.[7] LAT has an advantage of higher specificity than the in vivo tests, and is more practical. However, neither LAT nor the LTT is sensitive; this problem can be addressed by combining such tests with assays of drug-specific cytokine levels.[713] We performed LAT only.
The association between the HLA-B*58:01 allele and allopurinol-induced SCARs is well known, but the underlying mechanism remains unclear. We found that one patient reacted to allopurinol upon LAT. HLA-B genotyping would aid in prevention of allopurinol-induced SCARs, and LAT can serve to identify the causative agent. Further development of diagnostic methods is needed to provide a cost-effective, rapid, and accurate method predictive of the development of allopurinol-induced SCARs.
Figures and Tables
Figure 1
Patient 3: Erythematous macules with edema on the face (A); erythematous macules on the upper and lower extremities (B-C); and lymphocyte infiltration evident on skin biopsy (D-E).
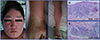
Figure 2
An allopurinol-positive lymphocyte activation test (stimulation index >2). CD69 was upregulated by allopurinol in CD4+ T cells from Patient 3 at both 48 and 72 h after culture. Sequence: negative control; various drug concentrations, and the phytohemagglutinin A (PHA) positive control.

Table 1
Clinical and laboratory characteristics of patients with allopurinol-induced SCARs (all had DRESS syndrome)

Acknowledgements
This research was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI15C1537).
References
1. Pirmohamed M, Friedmann PS, Molokhia M, Loke YK, Smith C, Phillips E, et al. Phenotype standardization for immune-mediated drug-induced skin injury. Clin Pharmacol Ther. 2011; 89:896–901. DOI: 10.1038/clpt.2011.79.

2. Mallal S, Nolan D, Witt C, Masel G, Martin AM, Moore C, et al. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet. 2002; 359:727–732.

3. Chung WH, Hung SI, Hong HS, Hsih MS, Yang LC, Ho HC, et al. Medical genetics: A marker for Stevens-Johnson syndrome. Nature. 2004; 428:486.
4. Hung SI, Chung WH, Liou LB, Chu CC, Lin M, Huang HP, et al. HLA-B*5801 allele as a genetic marker for severe cutaneous adverse reactions caused by allopurinol. Proc Natl Acad Sci USA. 2005; 102:4134–4139.

5. Kaniwa N, Saito Y, Aihara M, Matsunaga K, Tohkin M, Kurose K, et al. HLA-B locus in Japanese patients with anti-epileptics and allopurinol-related Stevens-Johnson syndrome and toxic epidermal necrolysis. Pharmacogenomics. 2008; 9:1617–1622. DOI: 10.2217/14622416.9.11.1617.

6. Kang HR, Jee YK, Kim YS, Lee CH, Jung JW, Kim SH, et al. Positive and negative associations of HLA class I alleles with allopurinol-induced SCARs in Koreans. Pharmacogenet Genomics. 2011; 21:303–307. DOI: 10.1097/FPC.0b013e32834282b8.

7. Romano A, Torres MJ, Castells M, Sanz ML, Blanca M. Diagnosis and management of drug hypersensitivity reactions. J Allergy Clin Immunol. 2011; 127:S67–S73. DOI: 10.1016/j.jaci.2010.11.047.

8. Kim MH, Shim EJ, Jung JW, Sohn SW, Kang HR. A case of allopurinol-induced fixed drug eruption confirmed with a lymphocyte transformation test. Allergy Asthma Immunol Res. 2012; 4:309–310. DOI: 10.4168/aair.2012.4.5.309.

9. Beeler A, Zaccaria L, Kawabata T, Gerber BO, Pichler WJ. CD69 upregulation on T cells as an in vitro marker for delayed-type drug hypersensitivity. Allergy. 2008; 63:181–188.

10. Ng CY, Yeh YT, Wang CW, Hung SI, Yang CH, Chang YC, et al. Impact of the HLA-B(*)58:01 allele and renal impairment on allopurinol-induced cutaneous adverse reactions. J Invest Dermatol. 2016; 136:1373–1381. DOI: 10.1016/j.jid.2016.02.808.

11. Jutkowitz E, Dubreuil M, Lu N, Kuntz KM, Choi HK. The cost-effectiveness of HLA-B*5801 screening to guide initial urate-lowering therapy for gout in the United States. Semin Arthritis Rheum. 2017; 46:594–600. DOI: 10.1016/j.semarthrit.2016.10.009.

12. In JW, Roh EY, Oh S, Shin S, Park KU, Song EY. Allele and haplotype frequencies of human leukocyte antigen-A, -B, -C, -DRB1, and -DQB1 from sequence-based DNA typing data in Koreans. Ann Lab Med. 2015; 35:429–435. DOI: 10.3343/alm.2015.35.4.429.

13. Lochmatter P, Beeler A, Kawabata TT, Gerber BO, Pichler WJ. Drug-specific in vitro release of IL-2, IL-5, IL-13, and IFN-gamma in patients with delayed-type drug hypersensitivity. Allergy. 2009; 64:1269–1278. DOI: 10.1111/j.1398-9995.2009.01985.x.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download