Abstract
Purpose
Under different culture conditions, periodontal ligament (PDL) stem cells are capable of differentiating into cementoblast-like cells, adipocytes, and collagen-forming cells. Several previous studies reported that because of the stem cells in the PDL, the PDL have a regenerative capacity which, when appropriately triggered, participates in restoring connective tissues and mineralized tissues. Therefore, this study analyzed the genes involved in mineralization during differentiation of human PDL (hPDL) cells, and searched for candidate genes possibly associated with the mineralization of hPDL cells.
Methods
To analyze the gene expression pattern of hPDL cells during differentiation, the hPDL cells were cultured in two conditions, with or without osteogenic cocktails (β-glycerophosphate, ascorbic acid and dexamethasone), and a DNA microarray analysis of the cells cultured on days 7 and 14 was performed. Reverse transcription-polymerase chain reaction was performed to validate the DNA microarray data.
Results
The up-regulated genes on day 7 by hPDL cells cultured in osteogenic medium were thought to be associated with calcium/iron/metal ion binding or homeostasis (PDE1A, HFE and PCDH9) and cell viability (PCDH9), and the down-regulated genes were thought to be associated with proliferation (PHGDH and PSAT1). Also, the up-regulated genes on day 14 by hPDL cells cultured in osteogenic medium were thought to be associated with apoptosis, angiogenesis (ANGPTL4 and FOXO1A), and adipogenesis (ANGPTL4 and SEC14L2), and the down-regulated genes were thought to be associated with cell migration (SLC16A4).
Conclusions
This study suggests that when appropriately triggered, the stem cells in the hPDL differentiate into osteoblasts/cementoblasts, and the genes related to calcium binding (PDE1A and PCDH9), which were strongly expressed at the stage of matrix maturation, may be associated with differentiation of the hPDL cells into osteoblasts/cementoblasts.
The periodontal ligament (PDL) is a fibrous connective tissue that surrounds and supports the tooth, and is located between the cementum of the root and the inner wall of the alveolar bone socket. It plays a crucial role in the maintenance and regeneration of periodontal tissue.
PDL contains a heterogeneous cell population at various stages of differentiation and lineage responsibility. Previous studies have suggested that PDL cells can form mineralized nodules in vitro and express bone-related proteins, such as alkaline phosphatase, bone sialoprotein and osteocalcin [1-4]. Recently, Seo et al. [5] reported that human PDL (hPDL) cells contain stem cells with a high proliferative capacity, self-renewal properties, multilineage differentiation potential and the capability of forming cementum/periodontal ligament-like tissue in vivo. Also, under different culture conditions, they suggested that isolated PDL stem cells are capable of differentiating into cementoblast-like cells, adipocytes, and collagen-forming cells. Particularly, cementoblastic/osteoblastic differentiation for the formation of new cementum or bone is very important to regeneration of the periodontium [6,7]. Commonly, osteoblasts form bone-like mineralized nodules in culture by undergoing three stages of differentiation: proliferation, extracellular matrix maturation and mineralization [8]. However, the exact molecular mechanism controlling the differentiation mechanism of progenitors in hPDL cells remain largely unknown.
The microarray has been successful in identifying differentiation stage-specific gene expression. Kulterer et al. [8] identified three novel candidate genes (ID4, CRYAB and SORT1) using microarray analysis of mesenchymal stem cell during osteogenic differentiation. Yamada et al. [9] reported that PLAP-1 regulated PDL cell mineralization negatively using a DNA microarray. Using a microarray, Lallier and Spencer [10] reported that PDL cells in culture are regulated by several factors that differentially stimulate a mineralized fate, a non-mineralized fate, or the propagation of a more naive phenotype. In a previous study using a cDNA microarray, Shin et al. [11] reported that at the stage of mineralization of hPDL cells, apoptosis-inducing agents were up-regulated, and anti-apoptosis activators were down-regulated. Even though there are many studies about the differentiation and mineralization of PDL, the analysis of the gene expressed in each stage during differentiation are not well understood and remains an active area of investigation [9-11]. Thus, the analysis of genes related to each stage of differentiation is very important.
Therefore, in the present study, using a culture system that facilitates the formations of mineralized nodules in hPDL cells, this study analyzed gene-expression profiles on days 7 and 14 of culture using a DNA microarray and sought candidate genes possibly associated with mineralization in an established line of hPDL cells characterized by the ability to differentiate into osteoblastic/cementoblastic lineages.
After receiving patient written consent, PDL tissues were obtained from the PDL of premolar teeth extracted during orthodontic treatment. Premolars (n=5) were collected from 5 female individuals aged 10 to 20 years, at Kyungpook National University Dental Hospital, Department of Periodontology.
The primary culture of the PDL cells was prepared using an explant technique according to a previous study [11]. The PDL cells were incubated in culture medium (Dulbecco's modified eagle medium [DMEM] with 10% fetal bovine serum [FBS], 100 U/mL penicillin and 100 µg/mL streptomycin) in a humidified atmosphere of 95% air. The medium was changed three times a week, and the hPDL cells used in this study were at 2 to 5 passages.
Mineralized nodule formation was evaluated by Dahl's method for calcium [12]. The hPDL cells were placed in a 24-well plate at a density of 2×104 cells/well and cultured in osteogenic medium (DMEM with 10% FBS, 50 µg/mL ascorbic acid, 10 mM β-glycerophospahte and 100 nM dexamethasone) or non-osteogenic medium (DMEM with 10% FBS) for 2 weeks. The cells were fixed in 2% paraformaldehyde neutral buffer solution and then stained with alizarin red S.
DNA microarray technologies were used to identify the differentiation-related genes of the hPDL cells. Five independently isolated hPDL cell samples were placed in a 6-well plate at a density of 1×105 cells/well. Culture plates were divided into 2 groups and cells were cultured in 1) osteogenic medium or 2) non-osteogenic medium for 7 and 14 days. On day 7 and 14, the total RNA was isolated from cultured cells using an RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany). The purity and integrity of the total RNA were verified using a UV spectrometer (Beckman DU530, Harlow Scientific, Arlington, MA, USA), respectively. Total RNA samples from different individuals were pooled and then the next experiments were carried out. Probe synthesis from total RNA samples, hybridization, detection, and scanning were performed according to standard protocols from Affymetrix Inc. (Santa Clara, CA, USA). Samples were analyzed by the use of the Affymetrix HG-U133 Plus 2.0 genechip, which contains 47,000 transcripts.
Expression profiles were analyzed using GeneChip Operating Software (GCOS, Affymetrix Inc.) and other programs. The Affymetrix software was used to identify changes in expression levels between the osteogenic medium group and non-osteogenic medium group using standard protocols recommended by Affymetrix. All genes with normalized data values >1.0 fold higher (up/down-regulation) than in the other groups were selected. Next, the genes with more than a 3-fold change and a signal value of 100 were filtered. Gene function analysis was performed by using the gene ontology mining tool of NetAffx, which is based on the Gene Ontology database (http://www.geneontology.org). Specification of the many gene annotations was also supplemented by further online database searches.
To validate the DNA microrarray data, some genes expressed at each function's groups on indicated days were selected, and their expression was measured by reverse transcription-polymerase chain reaction (RT-PCR).
The DNA was synthesized from the same total RNA as used for the DNA microarray. The oligonucleotide RT-PCR primers listed in Table 1 were purchased from Bioneer Corporation (Daegeon, Korea). Amplifications were performed in a PTC-2000 thermal cycler (MJ Research, Waltham, MA, USA) for 27 to 30 cycles after an initial 30 seconds denaturation at 94℃, annealed for 30 seconds at 56℃, and extended for 30 seconds at 72℃ in all primers. The amplification reaction products were resolved on 1% agarose/Tris-acetate-EDTA gels (Combrex BioScience, Rockland, ME, USA), electrophoresed at 100 mV, and visualized by ethidium-bromide staining.
The degree of mineralization in the hPDL cells cultured for 2 weeks by alizarin red S stain was assessed (Fig. 1). The hPDL cells cultured in non-osteogenic medium were not observed to have a mineralized nodule. However, the hPDL cells cultured in osteogenic medium were observed to have a mineralized nodule on day 14, but on day 7, a mineralized nodule was not observed in the osteogenic medium.
The five hPDL cell samples were cultured from each of the two different culture media from each day 7 and 14 and total RNA was isolated for analysis on the DNA microarray. An array of differentiation-related genes in the hPDL cells undergoing osteogenic differentiation was analyzed.
Among more than 47,000 genes analyzed, 111 genes were up-regulated more than 3-fold on day 7 in cells that were cultured in osteogenic medium, and 19 genes were down-regulated more than 3-fold in the osteogenic medium. On day 14, 77 genes were up-regulated, and 19 genes were down-regulated in the osteogenic medium.
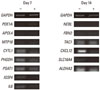
To validate the DNA microarray data, some genes expressed in each function's groups on the indicated days were selected, and their expression was measured by RT-PCR (Table 1). Twelve out of 14 genes were comparable with the DNA microarray data, suggesting that our microarray experiment was properly performed. TAC1 were expressed not significantly, and NEBL were not expressed (Fig. 2). Except for 2 genes, all genes were comparable with the DNA microarray data. A large part of the selected genes on indicated days were also expressed in other conditions.
By comparing the lists of differentially expressed genes from each of the two groups, a group of functionally expressed genes (Tables 2, 3, 4, 5) was classified. Gene function analysis was carried out by searching the National Center for Biotechnology Information database. The classified genes were related to signal transduction, transport, metabolism, cell adhesion, apoptosis, development, cell-cell signaling and angiogenesis during differentiation of hPDL cells.
On day 7, the up-regulated genes related to signal transduction were PDE1A, MYO10, APOB, and so on, and down-regulated genes were CYTL1 and IL6. On day 14, the up-regulated genes were ANGPTL4, EPHA3, RASL10B, etc., and the down-regulated genes were CXCL12, CCL8 and VLDLR. The genes up-regulated on both days were LEP, RAC3, and RASL11B, and those down-regulated both days were MX1 and WISP2. Notably, up- and down-regulated genes PDE1A, APOB, ALCAM, PSCD4, CYTL1, and IL6 were more strongly expressed on day 7 than on day 14, while ANGPTL4, EPHA3, RAB12, CXCL12, CCL8, and VLDLR were more strongly expressed on day 14 than on day 7.
On day 7, the up-regulated genes related to transport were APOL4, APOB, CLCN5, etc., and the down-regulated genes were PHGDH and SLC7A5. On day 14, the up-regulated genes were NEBL, SEC14L2, CLIC6, etc., and the down-regulated genes were SLC16A4, CCL8, and VLDLR. The genes up-regulated on both days were FABP4, STEAP4, APOD, etc., and the one down-regulated on both days was NPTX1. Interestingly, the up and down regulated genes APOB, CLCN5, HFE, SLC24A3, KCNK6, PHGDH and SLC7A5 were more strongly expressed on day 7 than on day 14, while NEBL, SEC14L2, CLIC6, RAB12, SLC16A4, CCL8, and VLDLR were more strongly expressed on day 14 than on day 7.
On day 7, the up-regulated genes related to metabolism were APOL4, EHHADH, APOB, etc., and the down-regulated genes were PSAT1, PHGDH, SLC7A5, and ASNS. On day 14, the up-regulated genes were ANGPTL4, PDK4, GNLY, etc., and the down-regulated genes were ALDH1A3 and VLDLR. The genes up-regulated on both days were CORIN, LEP, GGTLA1, etc., and the one down-regulated on both days was PTGS2. Notably, up- and down-regulated genes APOB, PDK3, CPM, MVK, PSAT1, PHGDH, SLC7A5, and ASNS were more strongly expressed on day 7 than on day 14, and ANGPTL4, PDK4, GNLY, ALDH1A3, while VLDLR were more strongly expressed on day 14 than on day 7.
On day 7, the up-regulated genes related to cell adhesion were PCDH9, NRP2, and ALCAM, and the down-regulated gene was IGSF4. On day 14, the up-regulated genes were FBN2 and PKP3, and the down-regulated gene was CXCL12. Of those related to cell adhesion on both days, the up-regulated genes were SAA1, COMP4, and ITGA10, and the down-regulated genes were VCAM1, CLDN1, and WISP2. Interestingly, up- and down-regulated genes such as PCDH9, NRP2, ALCAM, and IGSF4 were more strongly expressed on day 7 than day 14, while FBN2 and CXCL12 were more strongly expressed on day 14 than on day 7.
On day 7, the up-regulated gene related to apoptosis was MTP18, and the down-regulated gene was IL6. On day 14, up-regulated genes were ANGPTL4, FOXO1A and SEC14L2, while the down-regulated gene was ALDH1A3. For both days, the up-regulated gene was PLZF, and the down-regulated gene was MX1. Notably, the up- and down-regulated gene IL6 was more strongly expressed on day 7 than on day 14, while ANGPTL4, FOXO1A, SEC14L2 and ALDH1A3 were more strongly expressed on day 14 than on day 7.
On day 7, the up-regulated genes related to development were NRP2, SHC3, RNF103, etc., and the down-regulated gene was PHGDH. On day 14, the up-regulated genes were ANGPTL4, FOXO1A, LY6H, etc., and the down-regulated gene was ALDH1A3. For both days, up-regulated genes were PLZF, INS, GPM6B, COMP4, and FOXC2, and the down-regulated genes were NPTX1 and MEST. Notably, up- and down-regulated genes NRP2, SHC3, PHC2, and PHGDH were more strongly expressed on day 7 than on day 14, and ANGPTL4, FOXO1A, FRZB, and HCK were more strongly expressed on day 14 than on day 7.
On day 7, no up-regulated genes related to cell-cell signaling were observed, while the down-regulated gene was IL6. On day 14, the up-regulated genes were TAC1 and ADRA1B, and the down-regulated genes were CXCL12 and CCL8. For both days, up-regulated gene was LEP, and the down-regulated gene was WISP2. Interestingly, up- and down-regulated genes the gene IL6 was more strongly expressed on day 7 than on day 14, and TAC1, ADRA1B, CXCL12, and CCL8 were strongly expressed on day 14 than on day 7.
On day 7, the up-regulated gene related to angiogenesis was NRP2. On day 14, the up-regulated genes were ANGPTL4 and TNFAIP2. For both days, the up-regulated gene was FOXC2. However, on the indicated day, genes related to angiogenesis were not observed among the down-regulated genes. Nevertheless, up- and down-regulated genes the gene NRP2 was more strongly expressed on day 7 than on day 14, and ANGPTL4 was more strongly expressed on day 14 than on day 7.
PDL cells have been known as multipotential cells, so they have a capacity to differentiate into osteoblasts, cementoblasts, adipocytes, and collagen-forming cells. Several previous studies [5,13,14] have reported that because of the stem cells in the PDL, the PDL has a regenerative capacity to restore connective tissues and mineralized tissues when appropriately triggered. PDL cells have been reported to form mineralized nodules in vitro [5,15,16]. Osteoblasts form bone-like mineralized nodules by undergoing three stages of differentiation: proliferation, extracellular matrix maturation and mineralization. Actually, the stage of matrix maturation is around day 7, and the early stage of mineralization or late stage of matrix maturation is around day 14. Therefore the difference in gene expression in the cells cultured on days 7 and 14 was analyzed.
DNA microarray analysis of the cells cultured on days 7 and 14 under the osteogenic or non-osteogenic medium, was performed. The genes which were expressed in hPDL cells were classified by function. Each group was divided into the functions of signal transduction, transport, metabolism, cell adhesion, apoptosis, development, cell-cell signaling, and angiogenesis. To validate the DNA microarray data, some genes expressed at each function's groups on the indicated days were selected. The 12 out of 14 genes were comparable with DNA microarray data. Therefore, it was thought that the DNA microarray analysis is a reliable method for examining a gene expression profile.
It was considered that up or down regulated genes that were expressed more or less on day 7 than on day 14 were more important and related to each stage of differentiation of hPDL cells than other genes. Thus, focus was directed toward the genes which showed differences greater than 2-fold between day 7 and day 14.
On day 7, PDE1A, PSCD4, APOB, HFE, SLC24A3, KCNK6, CPM, MVK, PCDH9, NRP2, and SHC3 were more strongly up-regulated than on day 14 in osteogenic medium. The functions of these genes are related to calcium/iron/metal ion binding or homeostasis (PDE1A, HFE, SLC24A3, KCNK6, CPM, and PCDH9), cholesterol synthesis or homeostasis (APOB and MVK) and cell survival, migration, and invasion (NRP2). Genes PDE1A, HFE, PCDH9, and NRP2 were shown to have more than a 2-fold difference between day 7 and 14.
As members of the PDE1 family, PDE1A (phosphodiesterase 1A, calmodulin-dependent) is a Ca(2+)/calmodulin dependent PDEs that is activated by calmodulin in the presence of Ca(2+) [17,18]. It was reported that calmodulin regulates osteoblast differentiation [19]. Therefore, PDE1A is related to calmodulin binding, signal transduction and ion binding.
The protein encoded by HFE is a membrane protein that is similar to MHC class I-type proteins and associates with β2-microglobulin (β2M). It is thought that the protein encoded by HFE functions to regulate iron absorption by regulating the interaction of the transferrin receptor with transferrin.
PCDH9 (protocadherin 9) belongs to the protocadherin gene family, a subfamily of the cadherin superfamily. Cadherins are a superfamily of calcium-dependent adhesion molecules that play multiple roles in morphogenesis, including proliferation, migration, differentiation and cell-cell recognition [20]. It is thought that PCDH9 is related to calcium ion binding, protein binding and cell adhesion.
NRP2 (neuropilin 2) encodes a member of the neuropilin family of receptor proteins. NRP2 is related to protein binding, cell adhesion, cell differentiation, neural crest cell migration, and heart/nervous system/multicellular organismal development. Some studies have reported that NRP2 expression enhances cell survival, migration, and invasion [21].
Thus these findings suggested that related genes during the stage of matrix maturation of hPDL cells cultured in osteogenic medium were associated with calcium/iron/metal ion binding or homeostasis, and cell viability.
On day 14, ANGPTL4, EPHA3, RAB12, NEBL, SEC14L2, CLIC6, PDK4, GNLY, FBN2, FOXO1A, FRZB, HCK, TAC1, and ADRA1B were more strongly up-regulated than on day 7 in osteogenic medium. The functions of these genes are related to apoptosis and angiogenesis (ANGPTL4 and FOXO1A), adipogenesis (ANGPTL4, SEC14L2, and PDK4), calcium-binding or ion channel activity (FBN2 and CLIC6), migration (HCK and ADRA1B), and Wnt antagonist and regulation of bone development (FRZB). Interestingly, the genes ANGPTL4, SEC14L2, PDK4, and FOXO1A were shown to have more than a 2-fold difference between day 7 and day 14.
ANGPTL4 (angiopoietin-like 4) is a member of the angiopoietin/angiopoietin-like gene family and encodes a glycosylated, secreted protein with a fibrinogen C-terminal domain. Hopwood et al. [22] reported that adipogenesis and lipid and/or glucose metabolism related gene, ANGPTL4, was expressed in fractured human bone. Also, this gene was related to cell differentiation, multicellular organismal development, negative regulation of apoptosis, positive regulation of angiogenesis, and signal transduction.
SEC14L2 (SEC14-like 2) encodes a cytosolic protein that belongs to a family of lipid-binding proteins including Sec14p, alpha-tocopherol transfer protein, and cellular retinol-binding protein. SEC14L2 was related to phospholipid binding, vitamin E binding and cholesterol synthesis.
FOXO1A (forkhead box O1) belongs to the forkhead family of transcription factors, which are characterized by a distinct forkhead domain. FOXO1A was related to anti-apoptosis, blood vessel development, and regulation of cell proliferation.
Thus, these findings suggested that related genes during the early stage of mineralization or the late stage of matrix maturation by hPDL cells cultured in osteogenic medium were more associated with apoptosis, angiogenesis, and adipogenesis.
CYTL1, IL6, PHGDH, SLC7A5, PSAT1, ASNS, and IGSF4 were more strongly up-regulated genes on day 7 than on day 14 in non-osteogenic medium. The functions of these genes were related to proliferation (CYTL1, PHGDH, PSAT1, SLC7A5, and IGSF4) and inflammation (CYTL1 and IL6). In particular, the genes PHGDH and PSAT1 were shown to have more than a 2-fold difference between day 7 and day 14.
PHGDH (phosphoglycerate dehydrogenase) is related to processes such as cell cycle process and cell development. Cho et al. [23] reported that the human PHGDH gene is regulated at the transcriptional level in a tissue and is dependent on the cellular proliferation status. Pollari et al. [24] reported that L-serine biosynthesis genes such as PHGDH and PSAT1 stimulate osteoclastogenesis and cancer cell proliferation. Thus, these genes were related to proliferation.
Thus, these findings suggested that related genes during the stage of matrix maturation of hPDL cells cultured in non-osteogenic medium were more associated with proliferation.
On day 14, CXCL12, CCL8, SLC16A4, and ALDH1A3 were more strongly up-regulated genes than on day 7 in non-osteogenic medium. The functions of these genes were related to inflammation or the immune response (CXCL12 and CCL8), detoxification (ALDH1A3), and cell migration (SLC16A4). Notably, a gene like SLC16A4 showed a difference more than 2-fold between day 7 and day 14.
SLC16A4 (solute carrier family 16, member 4 [monocarboxylic acid transporter 5]) is a member of the SLC16 family of solute transporters. Gallagher et al. [25] reported that the specific interaction of SLC16A4 with β1 integrin may regulate cell migration.
Thus, these findings suggested that related genes during the early stage of mineralization or the late stage of matrix maturation by hPDL cells cultured in non-osteogenic medium were more associated with cell migration.
Based on these results, it is suggested that several genes related to proliferation or migration were expressed when hPDL cells were cultured in non-osteogenic medium. Also, expression of the genes related to calcium/iron/metal ion binding or homeostasis and cell viability were increased at the stage of matrix maturation, and expression of the genes related to apoptosis, angiogenesis, and adipogenesis were increased at the early stage of mineralization or the late stage of matrix maturation when hPDL cells were cultured in osteogenic medium.
In conclusion, this study suggests that when appropriately triggered, the stem cells in the hPDL differentiate into osteoblasts/cementoblasts, and, the genes related to calcium binding such as PDE1A, PCDH9, which was strongly expressed at the stage of matrix maturation, may be associated with differentiation of the hPDL cells into osteoblasts/cementoblasts. Further studies are needed to examine the precise function of these genes.
Figures and Tables
Figure 1
Alizarin red S staining of the mineralized nodules cultured for 7 days (A) and 14 days (B) with osteogenic medium and non-osteogenic medium. Circular photographs show human periodontal ligament cells in the culture plate (24 wells). On day 7, mineralized nodules were not observed at all, but on day 14, mineralized nodules were only observed in the osteogenic group.

Figure 2
Expression on day 7 and 14 of selected genes by human periodontal ligament (hPDL) cells cultured in osteogenic or non-osteogeneic medium. The hPDL cells were cultured for 7 days in an osteogenic medium (+) or non-osteogenic medium (-). The 12 out of 14 genes were comparable with DNA microarray data. The expression patterns of tachykinin, precursor 1 were not significant, and nebulette were not expressed. Similar results were obtained in 2 separate experiments and representative data are shown. GAPDH: glyceraldehyde-3-phosphate dehydrogenase, PDE1A: phosphodiesterase 1A, calmodulin-dependent, APOL4: apolipoprotein L, 4, MTP18: mitochondrial protein 18 kDa, CYTL1: cytokine-like 1, PHGDH: phosphoglycerate dehydrogenase, PSAT1: phosphoserine aminotransferase 1, IGSF4: cell adhesion molecule 1, IL6: interleukin 6 (interferon, beta 2), NEBL: nebulette, FBN2: fibrillin 2, TAC1: tachykinin, precursor 1, CXCL12: chemokine (C-X-C motif ) ligand 12, SLC16A4: solute carrier family 16, member 4 (monocarboxylic acid transporter 5), ALDH1A3: aldehyde dehydrogenase 1 family, member A3.
ACKNOWLEDGMENTS
This work was supported by Korea Science and Engineering Foundation (KOSEF) grant funded by the Korean Government (MEST) (No. 2010-0020544). The authors report no conflicts of interest related to this study.
References
1. Lekic P, Rojas J, Birek C, Tenenbaum H, McCulloch CA. Phenotypic comparison of periodontal ligament cells in vivo and in vitro. J Periodontal Res. 2001. 36:71–79.

2. Ivanovski S, Haase HR, Bartold PM. Expression of bone matrix protein mRNAs by primary and cloned cultures of the regenerative phenotype of human periodontal fibroblasts. J Dent Res. 2001. 80:1665–1671.

3. Murakami Y, Kojima T, Nagasawa T, Kobayashi H, Ishikawa I. Novel isolation of alkaline phosphatase-positive subpopulation from periodontal ligament fibroblasts. J Periodontol. 2003. 74:780–786.

4. Wu L, Wei X, Ling J, Liu L, Liu S, Li M, et al. Early osteogenic differential protein profile detected by proteomic analysis in human periodontal ligament cells. J Periodontal Res. 2009. 44:645–656.

5. Seo BM, Miura M, Gronthos S, Bartold PM, Batouli S, Brahim J, et al. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. 2004. 364:149–155.

6. Flores MG, Yashiro R, Washio K, Yamato M, Okano T, Ishikawa I. Periodontal ligament cell sheet promotes periodontal regeneration in athymic rats. J Clin Periodontol. 2008. 35:1066–1072.

7. Iwata T, Yamato M, Tsuchioka H, Takagi R, Mukobata S, Washio K, et al. Periodontal regeneration with multi-layered periodontal ligament-derived cell sheets in a canine model. Biomaterials. 2009. 30:2716–2723.

8. Kulterer B, Friedl G, Jandrositz A, Sanchez-Cabo F, Prokesch A, Paar C, et al. Gene expression profiling of human mesenchymal stem cells derived from bone marrow during expansion and osteoblast differentiation. BMC Genomics. 2007. 8:70.

9. Yamada S, Ozawa Y, Tomoeda M, Matoba R, Matsubara K, Murakami S. Regulation of PLAP-1 expression in periodontal ligament cells. J Dent Res. 2006. 85:447–451.

10. Lallier TE, Spencer A. Use of microarrays to find novel regulators of periodontal ligament fibroblast differentiation. Cell Tissue Res. 2007. 327:93–109.

11. Shin JH, Park JW, Yeo SI, Noh WC, Kim MK, Kim JC, et al. Identification of matrix mineralization-related genes in human periodontal ligament cells using cDNA microarray. J Korean Acad Periodontol. 2007. 37:Suppl. 447–463.

12. Dahl LK. A simple and sensitive histochemical method for calcium. Proc Soc Exp Biol Med. 1952. 80:474–479.

13. Bartold PM, Shi S, Gronthos S. Stem cells and periodontal regeneration. Periodontol 2000. 2006. 40:164–172.

14. Nagatomo K, Komaki M, Sekiya I, Sakaguchi Y, Noguchi K, Oda S, et al. Stem cell properties of human periodontal ligament cells. J Periodontal Res. 2006. 41:303–310.

15. Kobayashi M, Takiguchi T, Suzuki R, Yamaguchi A, Deguchi K, Shionome M, et al. Recombinant human bone morphogenetic protein-2 stimulates osteoblastic differentiation in cells isolated from human periodontal ligament. J Dent Res. 1999. 78:1624–1633.

16. Kitagawa M, Tahara H, Kitagawa S, Oka H, Kudo Y, Sato S, et al. Characterization of established cementoblast-like cell lines from human cementum-lining cells in vitro and in vivo. Bone. 2006. 39:1035–1042.

17. Michibata H, Yanaka N, Kanoh Y, Okumura K, Omori K. Human Ca2+/calmodulin-dependent phosphodiesterase PDE1A: novel splice variants, their specific expression, genomic organization, and chromosomal localization. Biochim Biophys Acta. 2001. 1517:278–287.

18. Fidock M, Miller M, Lanfear J. Isolation and differential tissue distribution of two human cDNAs encoding PDE1 splice variants. Cell Signal. 2002. 14:53–60.

19. Zayzafoon M, Fulzele K, McDonald JM. Calmodulin and calmodulin-dependent kinase IIalpha regulate osteoblast differentiation by controlling c-fos expression. J Biol Chem. 2005. 280:7049–7059.

20. Etzrodt J, Krishna-K K, Redies C. Expression of classic cadherins and delta-protocadherins in the developing ferret retina. BMC Neurosci. 2009. 10:153.
21. Gray MJ, Van Buren G, Dallas NA, Xia L, Wang X, Yang AD, et al. Therapeutic targeting of neuropilin-2 on colorectal carcinoma cells implanted in the murine liver. J Natl Cancer Inst. 2008. 100:109–120.

22. Hopwood B, Tsykin A, Findlay DM, Fazzalari NL. Gene expression profile of the bone microenvironment in human fragility fracture bone. Bone. 2009. 44:87–101.

23. Cho HM, Jun DY, Bae MA, Ahn JD, Kim YH. Nucleotide sequence and differential expression of the human 3-phosphoglycerate dehydrogenase gene. Gene. 2000. 245:193–201.

24. Pollari S, Käkönen SM, Edgren H, Wolf M, Kohonen P, Sara H, et al. Enhanced serine production by bone metastatic breast cancer cells stimulates osteoclastogenesis. Breast Cancer Res Treat. 2011. 125:421–430.

25. Gallagher SM, Castorino JJ, Philp NJ. Interaction of monocarboxylate transporter 4 with beta1-integrin and its role in cell migration. Am J Physiol Cell Physiol. 2009. 296:C414–C421.




 PDF
PDF ePub
ePub Citation
Citation Print
Print







 XML Download
XML Download