Abstract
Objective
Uterine leiomyomas are the most common tumor of the uterus. But the molecular causes of uterine leiomyoma remain unclear. We conducted the current investigation in order to elucidate the molecular mechanisms in the development of uterine leiomyoma.
Methods
We employed a new and accurate reverse transcription-polymerase chain reaction (RT-PCR) method that involved annealing control primers (ACPs) to identify the genes that are differently expressed in uterine leiomyoma.
Results
Using 120 ACPs, we identified and sequenced 14 differently expressed genes (DEGs) in uterine leiomyoma compared with normal myometrium. Basic Local Alignment Search Tool (BLAST) searches were performed to examine the known functions of these genes associated with uterine leiomyoma. We confirmed differently expressed patterns in more cases using the RT-PCR method. We also detected two novel genes, Tenascin-X and Leukemia Inhibitory Factor Receptor (LIFR), which had not yet been reported to have any functions associated with uterine leiomyoma. RT-PCR confirmation shows that both of these two genes are down-regulated in uterine leiomyoma.
Conclusion
Our results suggest that Tenascin-X and LIFR may play a role in the development of uterine leiomyoma. Although further studies are required to establish the precise mechanisms with which these genes are involved in the genesis of uterine leiomyoma, the present research is significant in that it is the first study which detects down-regulated novel genes in uterine leiomyoma using the ACP system.
Uterine Leiomyomas, also known as fibroids or myomas, are the most common uterine tumors in women of reproductive age. Remarkably, the prevalence of these benign tumors in the female population is 20 to 30%. Considered to be a major public health concern, uterine leiomyomas cause a variety of problems for women including menorrhagia, infertility, urinary incontinence, constipation, and abdominal pain. The presence of uterine leiomyomas during pregnancy has been associated with spontaneous abortion, premature labor, and dystocia. Consequently, uterine leiomyomas are the most frequent indication for hysterectomy, accounting for approximately 30% of hysterectomies (or about 2,000,000 procedures) annually in the United States.1 Therefore, the development of medical therapy is the only hope for such patients. Alternative approaches for studying patients with leiomyoma in various ethnic groups have revealed that steroid and polypeptide growth factors may be involved in the development and growth of these tumors,2-4 and that chromosome abnormality occurs in 7q22 and 12q14-15.5-7 It is hoped that identifying those genes activated or repressed in leiomyomas will provide new insights into their mechanisms, and possibly new methods for therapy. But the difficulty of identifying a gene responsible for a specialized function during a certain biological stage often arises because the gene is expressed at low levels, whereas the bulk of mRNA within a cell is highly abundant transcripts.8 As a result, to screen for differentially expressed gene transcripts in low concentrations, PCR amplification is required. Such techniques include differential display polymerase chain reaction (DD-PCR), serial analysis of gene expression (SAGE), differential hybridization, subtractive hybridization, and microarray (DNA chip). However, they have high false positivity and low reproducibility.
In order to eliminate these problems, we used a new and accurate reverse transcription polymerase chain reaction (RT-PCR) method that involves annealing control primers (ACPs) to identify the genes that are differently expressed in uterine leiomyomas.
Among the patients who underwent a hysterectomy due to uterine leiomyoma in the Department of Obstetrics and Gynecology at Ewha Womans University Mokdong Hospital from January 2004 to December 2004, 8 cases were randomly selected for this study. The patients' age were 49-58 years and all of them were menopausal. None of the patients had received any hormonal therapy within the previous 6 months. No patients had adenomyosis or other co-existent disease, confirmed by pathology. All patients were informed that their consent was required before the study.
Total RNAs extracted from uterine leiomyoma and normal myometrium were used for the synthesis of first-strand cDNAs by reverse transcriptase. Reverse transcription was performed for 1.5 hrs at 42℃ in a final reaction volume of 20 µl containing 3 µg of the purified total RNA, 4 µl of 5× reaction buffer (Promega, Madison, WI, USA), 5 µl of dNTPs (each 2 mM), 2 µl of 10 µM dT -ACP1 (5'-CGTGAATGCTGCGACTACGA TIIIIIT(18)-3'), 0.5 µl of RNasin RNase Inhibitor (40 U/µl; Promega), and 1 µl of Moloney murine leukemia virus reverse transcriptase (200 U/µl; Promega). First-strand cDNAs were diluted by the addition of 80 µl of ultra-purified water for the GeneFishing™ PCR, or by the addition of 180 µl of ultra-purified water for the SAFMTM PCR, and stored at -20℃ until used.
Differentially expressed genes (DEG) in two randomly selected cases were screened using the ACP-based PCR method with 120 ACPs9 using the GeneFishing™ DEG kits (Seegene, Seoul, South Korea). Briefly, second-strand cDNA synthesis was conducted at 50℃ during one cycle of first-stage PCR in a final reaction volume of 20 µl containing 3-5 µl (about 50 ng) of diluted first-strand cDNA, 1 µl of dT-ACP2 (10 µM), 1 µl of 10 µM arbitrary ACP, and 10 µl of 2 Master Mix (Seegene). The PCR protocol for second-strand synthesis was one cycle at 94℃ for 1 min, followed by 50℃ for 3 min, and 72℃ for 1 min. After second-strand DNA synthesis was completed, the second-stage PCR amplification protocol was 40 cycles of 94℃ for 40 sec, followed by 65℃ for 40 sec, 72℃ for 40 sec, followed by a 5 min final extension at 72℃. The amplified PCR products were separated in 2% agarose gel stained with ethidium bromide.
Differentially expressed bands were extracted from the gel by using the GENCLEAN II Kit (Q-BIO gene, Carlsbad, CA, USA), and directly cloned into a TOPO TA cloning vector (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The cloned plasmids were sequenced with ABI PRISM® 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). The identity of each product was confirmed by sequence homology analysis using Basic Local Alignment Search Tool (BLAST).10
The differential expression of DEG was confirmed by RT-PCR using each gene specific primer pair. The first-strand cDNA was normalized by the human beta-actin gene. The normalized cDNA was used as a template. The PCR reaction was conducted in a final reaction volume of 20 µl containing 2-4 µl (about 50 ng) of diluted first-strand cDNA, 1 µl of primer 5 (10 µM), 1 µl of primer 3 (10 µM), and 10 µl of 2 Master Mix (Seegene). The PCR amplification protocol was an initial 3 min denaturation at 94℃, followed by 20-25 cycles of 94℃ for 40 sec, 60℃ for 40 sec, 72℃ for 40 sec, and a 5 min final extension at 72℃. The amplified PCR products were separated in 2% agarose gel stained with ethidium bromide.
One hundred-twenty ACPs were used in RT-PCR for each paired sample from either leiomyoma or matched myometrium. The 39 and 59 differentially displayed cDNA bands with a visible intensity were yielded in case I and II respectively. Of these bands, fourteen (DEG 1, 2, 8, 9, 11, 12, 17, 18, 34, 40, 42, 50, 58, 59) were commonly expressed in both case I and II. DEG 9, 11, 18, 34, 50 of them showed increased signal and DEG 1, 2, 8, 12, 17, 40, 42, 58, 59 showed decreased signal in leiomyoma (Fig. 1).
After the RT-PCR subtraction procedure described above, the fourteen amplified products were cloned and sequenced. As a result of BLAST searching for fourteen DEGs, two (DEG 1, 59) were genes unknown to have a function associated in uterine leiomyoma (Fig. 2).
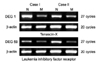
The differential expression of DEG 1, 59 were confirmed by RT-PCR using each gene specific primer pair. DEG 1 was identified as Tenascin-X, while DEG 59 was identified as leukemia inhibitory factor receptor (LIFR). All of them were down-regulated (Fig. 3). Two genes were confirmed in six more cases and showed the same results (Fig. 4).
Uterine leiomyoma is one of the most common benign tumors, occurring in 20% to 30% of women, accounting for significant morbidity and usually the need for major surgery. But women who need to preserve fertility or would like to avoid surgery require non-operative therapy. Unfortunately, there currently exists no medical therapy or prevention methods for these tumors. As a result, research for a new therapy is required. Molecular target therapy has been suggested as a possible avenue. The objective of this study was to serve as the molecular basis for the development of a new therapy.
Several previous studies, with these objectives, have been conducted. Up-regulated mRNAs in uterine leiomyoma were found by differential display of mRNA analysis. Its cDNA sequence was the same as that of the human S19 ribosomal protein.11 Four gene fragments (doublecortin, calpain 6, interleukin-17B, and proteolipid 1) were overexpressed in uterine leiomyoma compared with normal myometrium by microarray analysis.12 Although the DD-PCR method is simple, rapid, and only requires small amounts of total RNA, many investigators have experienced significantly high false-positive rates13 and poor reproducibility of results14 because of nonspecific annealing by short arbitrary primers.
To overcome these limitations, we used the annealing control primer (ACP) system which was recently developed. ACP™ technology uses primers that anneal specifically to the template and allow only genuine products to be amplified, a process that eliminates false-positive results.9 The basis of ACP™ technology is the unique tripartite structure of a specific oligonucleotide primer, which has 3'- and 5'-end distinct portions separated by a regulator, and the interaction of each portion of this primer during a two-stage PCR (Fig. 5).15 However, there are insufficient number of studies which have used the ACP system, so methodologic role of ACP systems for translational research has not been yet proven clearly.
In this study, we employed the ACP system and identified fourteen common differently expressed genes (DEGs) in 2 cases of leiomyoma. We excluded 12 of 14 genes those were known as non-specific and had known functions. The last 2, DEG 1 (Tenascin-X) and DEG 59 (Leukemia inhibitory factor receptor), were identified as novel genes by BLAST searching. Both were down regulated in leiomyoma.
Tenascins are a family of glycoproteins present in many extracellular matrix ECM) throughout the human body. Vertebrate genomes harbor four tenascin genes. The four tenascins have been labeled as tenascin-C, -R, -X, -W (Fig. 6). Tenascins are synthesized primarily by cells in connective tissues.16 Tenascin-X are prominent constituents of the muscle ECM secreted by fibroblasts present in the endomysium, perimysium, and epimysium. It is located in the central region of the human MHC III in a group with three other genes (RP, C4 and CYP21).
Tenascins contribute to matrix structure and collagen fiber stability and they are known to influence cell shape, migration and growth.17 Mice lacking tenascin-X display hyperelastic skin much like Ehler-Danlos patients with mutations in their tenascin-X gene. Tenascin-X null mice, generated by disruption of the Tenascin-X gene, display augmented invasion and metastasis of melanoma tumor cells due to increased activities of matrix metalloproteinases (MMP)-2 and MP-9. But the role of tenascin in the genesis of leiomyoma is not yet fully known.
Leukemia inhibitory factor (LIF) was first described as a factor that induces the differentiation of mouse myeloid leukemic M1 cells to macrophages.18 LIF is a member of the neuropoietic class of cytokines; this family includes interleukin (IL)-6, IL-11, ciliary neurotrophic factor (CNTF), cardiotrophin (CT-1), and oncostatin M (OSM).19 This factor is produced by a wide range of cells, including T-cells, thymic epithelial cells, astrocytes, neurons, mast cells, fibroblasts, keratinocytes, epithelial cells, endothelial cells, osteoblasts, synoviocytes, monocytes, and macrophages and secreted as a glycoprotein.20
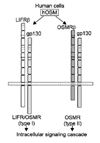
OSM is most closely related to LIF structurally, functionally, and genetically among the family members. OSM had long been considered as another LIF. In fact, these two cytokines act on a wide variety of cells and elicit diverse overlapping biological responses such as growth regulation, differentiation, gene expression, and cell survival in humans. The functional redundancy can be explained by their shared receptor subunit. OSM utilizes the LIFR in addition to its specific receptor (Fig. 7). OSM modulates growth of tumor and non-tumor cells either positively or negatively depending on the target cells. OSM inhibits the growth of several types of tumor cells such as solid tissue tumor cells, lung cancer cells, melanoma cells, breast cancer cells, and glioma cells. Besides tumor cells, OSM also inhibits the proliferation of normal mammary and breast epithelial cells. In contrast, OSM stimulates growth of AIDS-related Kaposi's sarcoma cells, myeloma cells, and plasmacytoma cells. OSM also stimulates the mitogenesis of normal dermal fibroblasts.21 However, there is no current data regarding leiomyoma.
In this study, Tenascin-X and LIFR were all down-regulated. We suggest that loss of Tenascin-X's function, which contribute to matrix structure and collagen fiber stability, may affect abnormal growth of uterine myometrium and that down-regulated LIFR may be associated with the development of uterine myoma since its ligand, OSM, is known to inhibit the growth of solid tumor cells. Therefore we can infer that there seems to be some role of these two genes in the genesis of uterine leiomyoma. Although further studies are required to establish the precise mechanisms, the current study is the first to have detected novel genes in leiomyoma using the ACP system. This serves as the basis for the development of a molecular target therapy in uterine leiomyoma.
Figures and Tables
References
1. Pokras R, Hufnagel VG. Hysterectomies in the United States 1965-84. 1987. Hyattsville, MD: US Department of Health and Human Services, Public Health Service, CDC;(Vital and health statistics; series 13, no. 92). DHHS publication no. (PHS) 88-1753.
2. Friedman AJ, Hoffman DI, Comite F, Browneller RW, Miller JD. Treatment of leiomyomata uteri with leuprolide acetate depot: A double-blind, placebo-controlled, multicenter study. The Leuprolide Study Group. Obstet Gynecol. 1991. 77:720–725.
3. Brandon DD, Erickson TE, Keenan EJ, Strawn EY, Novy MJ, Burry KA, et al. Estrogen receptor gene expression in human uterine leiomyomata. J Clin Endocrinol Metab. 1995. 80:1876–1881.
4. Yeh J, Rein M, Nowak R. Presence of messenger ribonucleic acid for epidermal growth factor (EGF) and EGF receptor demonstrable in monolayer cell cultures of myometria and leiomyomata. Fertil Steril. 1991. 56:997–1000.
5. Pandis N, Heim S, Bardi G, Flodérus UM, Willén H, Mandahl N, et al. Chromosome analysis of 96 uterine leiomyomas. Cancer Genet Cytogenet. 1991. 55:11–18.
6. Ishwad CS, Ferrell RE, Davare J, Meloni AM, Sandberg AA, Surti U. Molecular and cytogenetic analysis of chromosome 7 in uterine leiomyomas. Genes Chromosomes Cancer. 1995. 14:51–55.
7. Wanschura S, Belge G, Stenman G, Kools P, Dal Cin P, Schoenmakers E, et al. Mapping of the translocation breakpoints of primary pleomorphic adenomas and lipomas within a common region of chromosome 12. Cancer Genet Cytogenet. 1996. 86:39–45.
8. Wan JS, Sharp SJ, Poirier GM, Wagaman PC, Chambers J, Pyati J. Cloning differentially expressed mRNAs. Nat Biotechnol. 1996. 14:1685–1691.
9. Kim YJ, Kwak CI, Gu YY, Hwang IT, Chun JY. Annealing control primer system for identification of differentially expressed genes on agarose gels. Biotechniques. 2004. 36:424–434.
10. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990. 215:403–410.
11. Li B, Sun M, He B, Yu J, Zhang YD, Zhang YL. Identification of differentially expressed genes in human uterine leiomyomas using differential display. Cell Res. 2002. 12:39–45.
12. Skubitz KM, Skubitz AP. Differential gene expression in uterine leiomyoma. J Lab Clin Med. 2003. 141:297–308.
13. Debouck C. Differential display or differential dismay? Curr Opin Biotechnol. 1995. 6:597–599.
14. Liang P, Pardee AB. Recent advances in differential display. Curr Opin Immunol. 1995. 7:274–280.
15. Hwang KC, Cui XS, Park SP, Shin MR, Park SY, Kim EY, et al. Identification of differentially regulated genes in bovine blastocysts using an annealing control primer system. Mol Reprod Dev. 2004. 69:43–51.
16. Chiquet-Ehrismann R. Tenascins. Int J Biochem Cell Biol. 2004. 36:986–990.
17. Chiquet-Ehrismann R, Tucker RP. Connective tissues: Signalling by tenascins. Int J Biochem Cell Biol. 2004. 36:1085–1089.
18. Gearing DP, Gough NM, King JA, Hilton DJ, Nicola NA, Simpson RJ, et al. Molecular cloning and expression of cDNA encoding a murine myeloid leukemia inhibitory factor (LIF). EMBO. 1987. 6:3995–4002.
19. Turnley AM, Bartlett PF. Cytokines that signal through the leukemia inhibitory factor receptor-beta complex in the nervous system. J Neurochem. 2000. 74:889–899.
20. McKenzie RC, Szepietowski J. Cutaneous leukemia inhibitory factor and its potential role in the development of skin tumors. Dermatol Surg. 2004. 30:279–290.
21. Tanaka M, Miyajima A. Oncostatin M, a multifunctional cytokine. Rev Physiol Biochem Pharmacol. 2003. 149:39–52.




 PDF
PDF ePub
ePub Citation
Citation Print
Print







 XML Download
XML Download