Abstract
Background
Multilocus sequence typing (MLST) is useful in determining the long-term evolutionary process and minimizes differences in experimental results across individuals and laboratories. It is also useful in determining evolutionary origins and backgrounds of bacterial species. This study carries out MLST analysis on VanA-type vancomycin-resistant Enterococcus faecium isolated from patient specimens in a single university hospital over nine years in order to observe changes in genetic evolution over time.
Methods
During the years from 2007 to 2015, 44 clinical isolates of vanA-containing E. faecium were collected from Ajou University Hospital in Korea. Species were identified by the VitekII system (bio-Merieux, USA), and antibiotic susceptibility testing was performed by disk diffusion and E-test according to Clinical and Laboratory Standards Institute (CLSI) guidelines. To determine genetic relatedness, matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF M/S) was employed. To characterize clonal diversity, MLST analysis was used.
Results
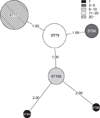
All isolates were highly resistant to ampicillin, ciprofloxacin, and vancomycin but showed variable levels of resistance to teicoplanin. The 44 clinical isolates were genetically unrelated according to MALDI-TOF M/S analysis. MLST showed that the clinical isolates harbored 6 sequence types (ST), with ST17 (n=19) being the most common, followed by ST78 (n=13), ST192 (n=6), ST64 (n=4), ST262 (n=1), and ST414 (n=1).
Conclusion
The MLST analysis showed that the sequence types of most isolates belonged to clonal complex 17 This is consistent with outbreaks in hospitals. We had single observations for ST262 and ST414, suggesting that they were random occurrences. MLST can be useful for speculating the genetic evolution of VanA-containing E. faecium isolates.
Figures and Tables
Fig. 1
Minimum spanning tree from 44 strains after MLST analysis. Thick solid lines indicate the single locus variation. Thin solid lines indicate the double locus variation.
References
1. Park JW, Kim YR, Shin WS, Kang MW, Han KJ, Shim SI. Susceptibility tests of vancomycin-resistant enterococci. Korean J Infect Dis. 1992; 24:133–137.
2. Boyd DA, Willey BM, Fawcett D, Gillani N, Mulvey MR. Molecular characterization of Enterococcus faecalis N06-0364 with low-level vancomycin resistance harboring a novel D-Ala-D-Ser gene cluster, vanL. Antimicrob Agents Chemother. 2008; 52:2667–2672.
3. McKessar SJ, Berry AM, Bell JM, Turnidge JD, Paton JC. Genetic characterization of vanG, a novel vancomycin resistance locus of Enterococcus faecalis. Antimicrob Agents Chemother. 2000; 44:3224–3228.
4. Xu X, Lin D, Yan G, Ye X, Wu S, Guo Y, et al. vanM, a new glycopeptide resistance gene cluster found in Enterococcus faecium. Antimicrob Agents Chemother. 2010; 54:4643–4647.
5. Lebreton F, Depardieu F, Bourdon N, Fines-Guyon M, Berger P, Camiade S, et al. D-Ala-d-Ser VanN-type transferable vancomycin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 2011; 55:4606–4612.
6. Lee WG, Jernigan JA, Rasheed JK, Anderson GJ, Tenover FC. Possible horizontal transfer of the vanB2 gene among genetically diverse strains of vancomycin-resistant Enterococcus faecium in a Korean hospital. J Clin Microbiol. 2001; 39:1165–1168.
7. Lee WG. Resistance mechanism and epidemiology of vancomycinresistant enterococci. Korean J Clin Microbiol. 2008; 11:71–77.
8. Homan WL, Tribe D, Poznanski S, Li M, Hogg G, Spalburg E, et al. Multilocus sequence typing scheme for Enterococcus faecium. J Clin Microbiol. 2002; 40:1963–1971.
9. Schlebusch S, Price GR, Gallagher RL, Horton-Szar V, Elbourne LD, Griffin P, et al. MALDI-TOF MS meets WGS in a VRE outbreak investigation. Eur J Clin Microbiol Infect Dis. 2017; 36:495–499.
10. Nowakiewicz A, Ziółkowska G, Zięba P, Gnat S, TroŚciańczyk A, Adaszek Ł. Characterization of multidrug resistant E. faecalis strains from pigs of local origin by ADSRRS-fingerprinting and MALDI-TOF MS; Evaluation of the compatibility of methods employed for multidrug resistance analysis. PLoS One. 2017; 12:e0171160.
11. McCarthy KL, Kidd TJ, Paterson DL. Molecular epidemiology of Pseudomonas aeruginosa bloodstream infection isolates in a non-outbreak setting. J Med Microbiol. 2017; 66:154–159.
12. Kainer MA, Devasia RA, Jones TF, Simmons BP, Melton K, Chow S, et al. Response to emerging infection leading to outbreak of linezolid-resistant enterococci. Emerg Infect Dis. 2007; 13:1024–1030.
13. Enright MC. Multilocus sequence typing. Trends Microbiol. 1999; 7:482–487.
14. Galloway-Peña JR, Nallapareddy SR, Arias CA, Eliopoulos GM, Murray BE. Analysis of clonality and antibiotic resistance among early clinical isolates of Enterococcus faecium in the United States. J Infect Dis. 2009; 200:1566–1573.
15. Gordts B, Van Landuyt H, Ieven M, Vandamme P, Goossens H. Vancomycin-resistant enterococci colonizing the intestinal tracts of hospitalized patients. J Clin Microbiol. 1995; 33:2842–2846.
16. Mato R, de Lencastre H, Roberts RB, Tomasz A. Multiplicity of genetic backgrounds among vancomycin-resistant Enterococcus faecium isolates recovered from an outbreak in a New York City hospital. Microb Drug Resist. 1996; 2:309–317.
17. Oh JY, Her SH, Seo SY, Lee YC, Lee JC, Kim J, et al. Antimicrobial resistance and multilocus sequence typing of vancomycin-resistant Enterococcus faecium isolated from clinical specimens. J Bacteriol Virol. 2008; 38:19–27.
18. Lee WG, Lee SM, Kim YS. Molecular characterization of Enterococcus faecium isolated from hospitalized patients in Korea. Lett Appl Microbiol. 2006; 43:274–279.
19. Yang JX, Li T, Ning YZ, Shao DH, Liu J, Wang SQ, et al. Molecular characterization of resistance, virulence and clonality in vancomycin-resistant Enterococcus faecium and Enterococcus faecalis: a hospital-based study in Beijing, China. Infect Genet Evol. 2015; 33:253–260.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download