Abstract
Background
To investigate the national molecular epidemiology and resistance profiles of vancomycin-intermediate Staphylococcus aureus (VISA), we analyzed the characteristics of methicillin-resistant Staphylococcus aureus (MRSA) collected from clinical samples at tertiary or general hospitals participating in a nationwide surveillance program for VISA and vancomycin-resistant Staphylococcus aureus (VRSA) in Korea during an 12-week period in each year from 2007 to 2013.
Methods
VISA was defined by agar dilution, broth dilution and E-test methods with vancomycin minimum inhibitory concentrations of >2 μ g/mL. All VISA isolates were characterized by multilocus sequence typing, staphylococcal cassette chromosome mec typing, spa typing, accessory gene regulator typing, Diversilab analysis, and antibiogram analysis.
Results
Of 109,345 MRSA isolates, 87,354 were screened and 426 isolates were identified as positive on brain heart infusion agar containing 4 μ g/mL vancomycin (BHI-V4). Of 426 isolates, 76 isolates were identified as VISA. No VRSA isolates were detected among the isolates. Overall, a total of 6 genotypes were identified among VISA strains and the predominant clones were ST5-II-t2460, ST72-IV-t324, and ST239-III-t037 (44.7%, 15.8%, and 10.5%, respectively). Of note, ST72-IV-t324 clones are known to be a typical community-associated MRSA. ST239-III-t037 strains were more resistant to trimethoprim-sulfa-methoxazole than any other type of strain. ST72-IV-t324 strains were susceptible to all of the antimicrobial agents tested except erythromycin and daptomycin. All of the VISA isolates were susceptible to linezolid and quinupristin-dalfopristin.
Go to : 
References
1. Howden BP, Peleg AY, Stinear TP. The evolution of vancomycin intermediate Staphylococcus aureus (VISA) and heterogenous-VISA. Infect Genet Evol. 2014; 21:575–82.

2. Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover FC. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997; 40:135–6.

3. Richter SS, Satola SW, Crispell EK, Heilmann KP, Dohrn CL, Riahi F, et al. Detection of Staphylococcus aureus isolates with heterogeneous intermediate-level resistance to vancomycin in the United States. J Clin Microbiol. 2011; 49:4203–7.

4. Rybak MJ, Leonard SN, Rossi KL, Cheung CM, Sader HS, Jones RN. Characterization of vancomycin-heteroresistant Staphylococcus aureus from the metropolitan area of Detroit, Michigan, over a 22-year period (1986 to 2007). J Clin Microbiol. 2008; 46:2950–4.
5. Adam HJ, Louie L, Watt C, Gravel D, Bryce E, Loeb M, et al. Detection and characterization of heterogeneous vancomycin-intermediate Staphylococcus aureus isolates in Canada: results from the Canadian Nosocomial Infection Surveillance Program, 1995–2006. Antimicrob Agents Chemother. 2010; 54:945–9.
6. Sun W, Chen H, Liu Y, Zhao C, Nichols WW, Chen M, et al. Prevalence and characterization of heterogeneous vancomycin-intermediate Staphylococcus aureus isolates from 14 cities in China. Antimicrob Agents Chemother. 2009; 53:3642–9.
7. Kirby A, Graham R, Williams NJ, Wootton M, Broughton CM, Alanazi M, et al. Staphylococcus aureus with reduced glyco-peptide susceptibility in Liverpool, UK. J Antimicrob Chemother. 2010; 65:721–4.

8. Wootton M, Howe RA, Hillman R, Walsh TR, Bennett PM, MacGowan AP. A modified population analysis profile (PAP) method to detect heteroresistance to vancomycin in Staphylococcus aureus in a UK hospital. J Antimicrob Chemother. 2001; 47:399–403.

9. Campanile F, Borbone S, Perez M, Bongiorno D, Cafiso V, Bertuccio T, et al. Heteroresistance to glycopeptides in Italian meticillin-resistant Staphylococcus aureus (MRSA) isolates. Int J Antimicrob Agents. 2010; 36:415–9.

10. Garnier F, Chainier D, Walsh T, Karlsson A, Bolmström A, Grelaud C, et al. A 1 year surveillance study of glycopeptide-intermediate Staphylococcus aureus strains in a French hospital. J Antimicrob Chemother. 2006; 57:146–9.

11. Bartley J. First case of VRSA identified in Michigan. Infect Control Hosp Epidemiol. 2002; 23:480.
12. Melo-Cristino J, Resina C, Manuel V, Lito L, Ramirez M. First case of infection with vancomycin-resistant Staphylococcus aureus in Europe. Lancet. 2013; 382:205.

13. Moravvej Z, Estaji F, Askari E, Solhjou K, Naderi Nasab M, Saadat S. Update on the global number of vancomycin-resistant Staphylococcus aureus (VRSA) strains. Int J Antimicrob Agents. 2013; 42:370–1.

14. Walters MS, Eggers P, Albrecht V, Travis T, Lonsway D, Hovan G, et al. Vancomycin-Resistant Staphylococcus aureus – Delaware, 2015. MMWR Morb Mortal Wkly Rep. 2015; 64:1056.
15. Tenover FC and Moellering RC Jr. The rationale for revising the Clinical and Laboratory Standards Institute vancomycin minimal inhibitory concentration interpretive criteria for Staphylococcus aureus. Clin Infect Dis. 2007; 44:1208–15.
16. van Hal SJ, Lodise TP, Paterson DL. The clinical significance of vancomycin minimum inhibitory concentration in Staphylococcus aureus infections: a systematic review and metaanalysis. Clin Infect Dis. 2012; 54:755–71.

17. Chung G, Cha J, Han S, Jang H, Lee K, Yoo J, et al. Nationwide surveillance study of vancomycin intermediate Staphylococcus aureus strains in Korean hospitals from 2001 to 2006. J Microbiol Biotechnol. 2010; 20:637–42.
18. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000; 38:1008–15.
19. Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003; 41:5442–8.
20. Koreen L, Ramaswamy SV, Graviss EA, Naidich S, Musser JM, Kreiswirth BN. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J Clin Microbiol. 2004; 42:792–9.
21. Oliveira DC and de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002; 46:2155–61.
22. Gilot P, Lina G, Cochard T, Poutrel B. Analysis of the genetic variability of genes encoding the RNA III-activating components Agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J Clin Microbiol. 2002; 40:4060–7.
23. Shutt CK, Pounder JI, Page SR, Schaecher BJ, Woods GL. Clinical evaluation of the DiversiLab microbial typing system using repetitive-sequence-based PCR for characterization of Staphylococcus aureus strains. J Clin Microbiol. 2005; 43:1187–92.
24. Chung DR, Lee C, Kang YR, Baek JY, Kim SH, Ha YE, et al. Genotype-specific prevalence of heterogeneous vancomycin-intermediate Staphylococcus aureus in Asian countries. Int J Antimicrob Agents. 2015; 46:338–41.

25. Zhang S, Sun X, Chang W, Dai Y, Ma X. Systematic review and metaanalysis of the epidemiology of vancomycin-intermediate and heterogeneous vancomycin-intermediate Staphylococcus aureus isolates. PLoS One. 2015; 10:e0136082.

26. Hu J, Ma XX, Tian Y, Pang L, Cui LZ, Shang H. Reduced vancomycin susceptibility found in methicillin-resistant and methicillin-sensitive Staphylococcus aureus clinical isolates in Northeast China. PLoS One. 2013; 8:e73300.

27. Liu C and Chambers HF. Staphylococcus aureus with heterogeneous resistance to vancomycin: epidemiology, clinical significance, and critical assessment of diagnostic methods. Antimicrob Agents Chemother. 2003; 47:3040–5.
28. Havaei SA, Azimian A, Fazeli H, Naderi M, Ghazvini K, Samiee SM, et al. Genetic characterization of methicillin resistant and sensitive, vancomycin intermediate Staphylococcus aureus strains isolated from different iranian hospitals. ISRN Microbiol. 2012; 2012:215275.

29. DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010; 375:1557–68.

30. Chuang YY and Huang YC. Molecular epidemiology of community-associated meticillin-resistant Staphylococcus aureus in Asia. Lancet Infect Dis. 2013; 13:698–708.
31. Park SH, Park C, Yoo JH, Choi SM, Choi JH, Shin HH, et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus strains as a cause of healthcare-associated bloodstream infections in Korea. Infect Control Hosp Epidemiol. 2009; 30:146–55.
32. Joo EJ, Choi JY, Chung DR, Song JH, Ko KS. Characteristics of the community-genotype sequence type 72 methicillin-resistant Staphylococcus aureus isolates that underlie their persistence in hospitals. J Microbiol. 2016; 54:445–50.

33. Prakash V, Lewis JS 2nd, Jorgensen JH. Vancomycin MICs for methicillin-resistant Staphylococcus aureus isolates differ based upon the susceptibility test method used. Antimicrob Agents Chemother. 2008; 52:4528.
34. Sader HS, Rhomberg PR, Jones RN. Nine-hospital study comparing broth microdilution and Etest method results for vancomycin and daptomycin against methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2009; 53:3162–5.
Go to : 
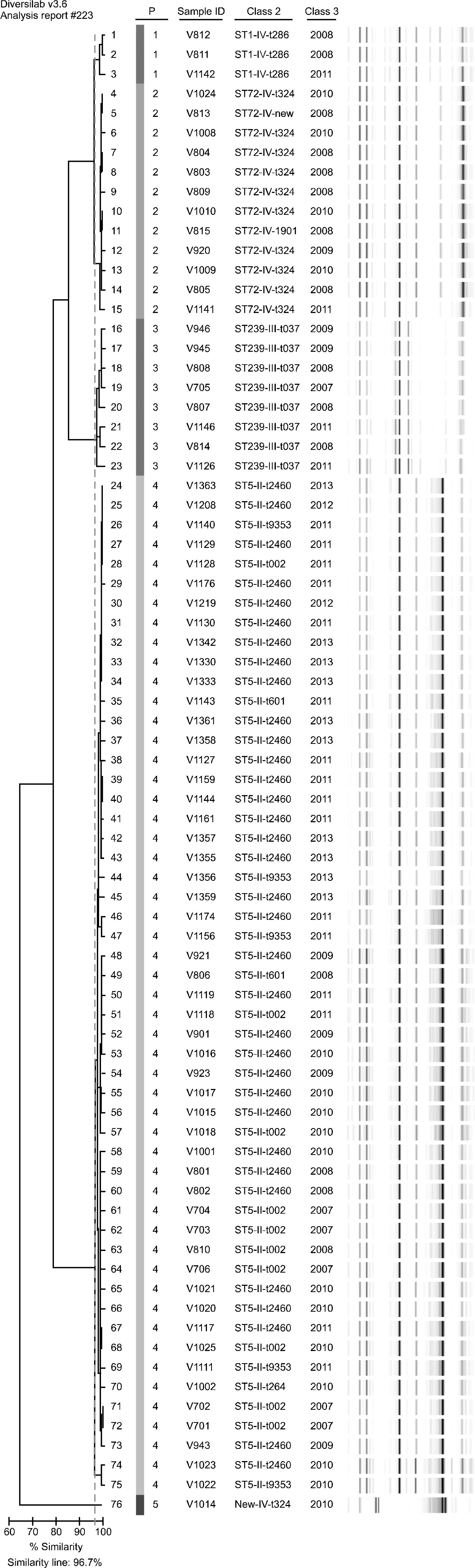 | Fig. 1.Cluster analysis and virtual gel image from Diversilab-generated fingerprints of the 76 VISA isolates, including corresponding typing data from molecular type (class 2) and year (class 3). Colored marks indicate the clonal clustering results. |
Table 1.
Screening of resistance to vancomycin of clinical MRSA isolates from 2007 to 2013 in Korea
Table 2.
Comparison of vancomycin MICs determined by broth microdilution, agar dilution, and E-test
Table 3.
Molecular characteristics of VISA isolates
Table 4.
Antimicrobial resistance profiles of VISA clones




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download