초록
Background
Vancomycin-resistant Enterococci (VRE) infections are caused by Enterococcus faecium in about 90% of the cases but can also be caused by Enterococcus faecalis. Thus, this study investigates factors that cause a low isolation rate of vancomy-cin-resistant E. faecalis (VREfs). To this end, the au-thors study the clinical traits, resistant gene structure, genomic classification, and molecular characteristics of the virulent factor.
Methods
From January 2001 through September 2011, 17 vanA-containing E. faecalis isolates were collected from hospitalized patients at Ajou University Hospital in Korea. Identification, antimicrobial susceptibility testing, and PCR of van and esp genes were performed. Pulsed-field gel electrophoresis (PFGE) was used for strain typing. PCR and sequencing of the internal regions of Tn1546 were performed for structural analysis of the van gene.
Results
Of 4,235 VRE infections, 3,918 (92.5%) were caused by E. faecium, and 95 (2.2%) were caused by E. faecalis. In 67% of VREfs infections, there was a preceding occurrence of E. faecium infection. All isolates were of genotype vanA. Our isolates were div-ided into three types according to the distribution of IS elements integrated into Tn1546 (types I to IIb). The PFGE results showed no clonal relatedness among isolates.
Go to : 
REFERENCES
1.Park JW., Kim YR., Shin WS., Kang MW., Han KJ., Shim SI. Susceptibility tests of vancomycin-resistant enterococci. Korean J Infect Dis. 1992. 24:133–7.
2.Boyd DA., Willey BM., Fawcett D., Gillani N., Mulvey MR. Molecular characterization of Enterococcus faecalis N06-0364 with low-level vancomycin resistance harboring a novel D-Ala-D-Ser gene cluster, vanL. Antimicrob Agents Chemother. 2008. 52:2667–72.
3.McKessar SJ., Berry AM., Bell JM., Turnidge JD., Paton JC. Genetic characterization of vanG, a novel vancomycin resistance locus of Enterococcus faecalis. Antimicrob Agents Chemother. 2000. 44:3224–8.
4.Xu X., Lin D., Yan G., Ye X., Wu S., Guo Y, et al. vanM, a new glycopeptide resistance gene cluster found in Enterococcus faecium. Antimicrob Agents Chemother. 2010. 54:4643–7.
5.Lebreton F., Depardieu F., Bourdon N., Fines-Guyon M., Berger P., Camiade S, et al. D-Ala-d-Ser VanN-type transferable vancomycin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 2011. 55:4606–12.
6.Toledo-Arana A., Valle J., Solano C., Arrizubieta MJ., Cucarella C., Lamata M, et al. The enterococcal surface protein, Esp, is in-volved in Enterococcus faecalis biofilm formation. Appl Environ Microbiol. 2001. 67:4538–45.
7.Deshpande LM., Fritsche TR., Moet GJ., Biedenbach DJ., Jones RN. Antimicrobial resistance and molecular epidemiology of vancomy-cin-resistant enterococci from North America and Europe: a report from the SENTRY antimicrobial surveillance program. Diagn Microbiol Infect Dis. 2007. 58:163–70.

8.Cockerill FR. Performance standards for antimicrobial susceptibility testing: Twenty-third informational supplement M100-S23. Wayne, PA: CLSI. 2013.
9.Willems RJ., Top J., van den Braak N., van Belkum A., Mevius DJ., Hendriks G, et al. Molecular diversity and evolutionary relation-ships of Tn1546-like elements in enterococci from humans and animals. Antimicrob Agents Chemother. 1999. 43:483–91.
10.Leavis H., Top J., Shankar N., Borgen K., Bonten M., van Embden J, et al. A novel putative enterococcal pathogenicity island linked to the esp virulence gene of Enterococcus faecium and associated with epidemicity. J Bacteriol. 2004. 186:672–82.
11.Murray BE., Singh KV., Heath JD., Sharma BR., Weinstock GM. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990. 28:2059–63.

12.Tenover FC., Weigel LM., Appelbaum PC., McDougal LK., Chaitram J., McAllister S, et al. Vancomycin-resistant Staphyloco-ccus aureus isolate from a patient in Pennsylvania. Antimicrob Agents Chemother. 2004. 48:275–80.
13.Arthur M., Molinas C., Depardieu F., Courvalin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993. 175:117–27.
14.Palepou MF., Adebiyi AM., Tremlett CH., Jensen LB., Woodford N. Molecular analysis of diverse elements mediating VanA glycopeptide resistance in enterococci. J Antimicrob Chemother. 1998. 42:605–12.

15.Darini AL., Palepou MF., Woodford N. Nucleotide sequence of IS1542, an insertion sequence identified within VanA glycopeptide resistance elements of enterococci. FEMS Microbiol Lett. 1999. 173:341–6.
16.Yu HS., Seol SY., Cho DT. Diversity of tn1546-like elements in vancomycin-resistant enterococci isolated from humans and poultry in Korea. J Clin Microbiol. 2003. 41:2641–3.
17.Lee WG., Huh JY., Cho SR., Lim YA. Reduction in glycopeptide resistance in vancomycin-resistant enterococci as a result of vanA cluster rearrangements. Antimicrob Agents Chemother. 2004. 48:1379–81.
18.Park IJ., Lee WG., Lim YA., Cho SR. Genetic rearrangements of TN1546-like elements in vancomycin-resistant Enterococcus faecium isolates collected from hospitalized patients over a seven-year period. J Clin Microbiol. 2007. 45:3903–8.
19.Sava IG., Heikens E., Kropec A., Theilacker C., Willems R., Huebner J. Enterococcal surface protein contributes to persistence in the host but is not a target of opsonic and protective antibodies in Enterococcus faecium infection. J Med Microbiol. 2010. 59:1001–4.
20.Tendolkar PM., Baghdayan AS., Gilmore MS., Shankar N. Enterococcal surface protein, Esp, enhances biofilm formation by Enterococcus faecalis. Infect Immun. 2004. 72:6032–9.
21.Heikens E., Bonten MJ., Willems RJ. Enterococcal surface protein Esp is important for biofilm formation of Enterococcus faecium E1162. J Bacteriol. 2007. 189:8233–40.
22.Shankar V., Baghdayan AS., Huycke MM., Lindahl G., Gilmore MS. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect Immun. 1999. 67:193–200.
23.Top J., Willems R., Bonten M. Emergence of CC17 Enterococcus faecium: from commensal to hospital-adapted pathogen. FEMS Immunol Med Microbiol. 2008. 52:297–308.
24.Comerlato CB., Resende MC., Caierão J., d'Azevedo PA. Presence of virulence factors in Enterococcus faecalis and Enterococcus faecium susceptible and resistant to vancomycin. Mem Inst Oswal-do Cruz. 2013. 108:590–5.
Go to : 
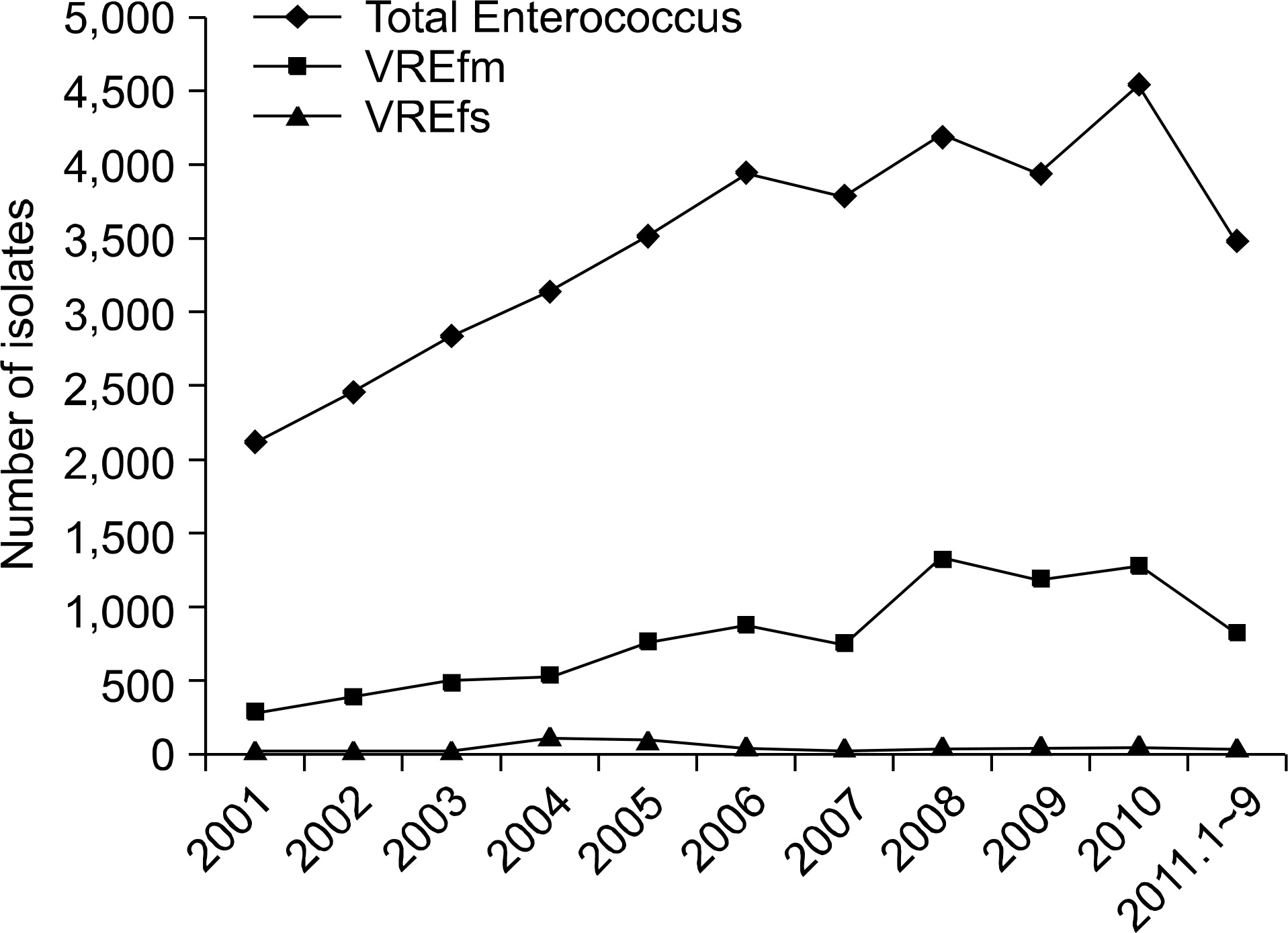 | Fig. 1.The number of isolated enterococcal species according to 11 years. Abbreviations: VREfm, vancomycin-resistant Enterococcus faecium; VREfs, vancomycin resistant-Enterococcus faecalis. |
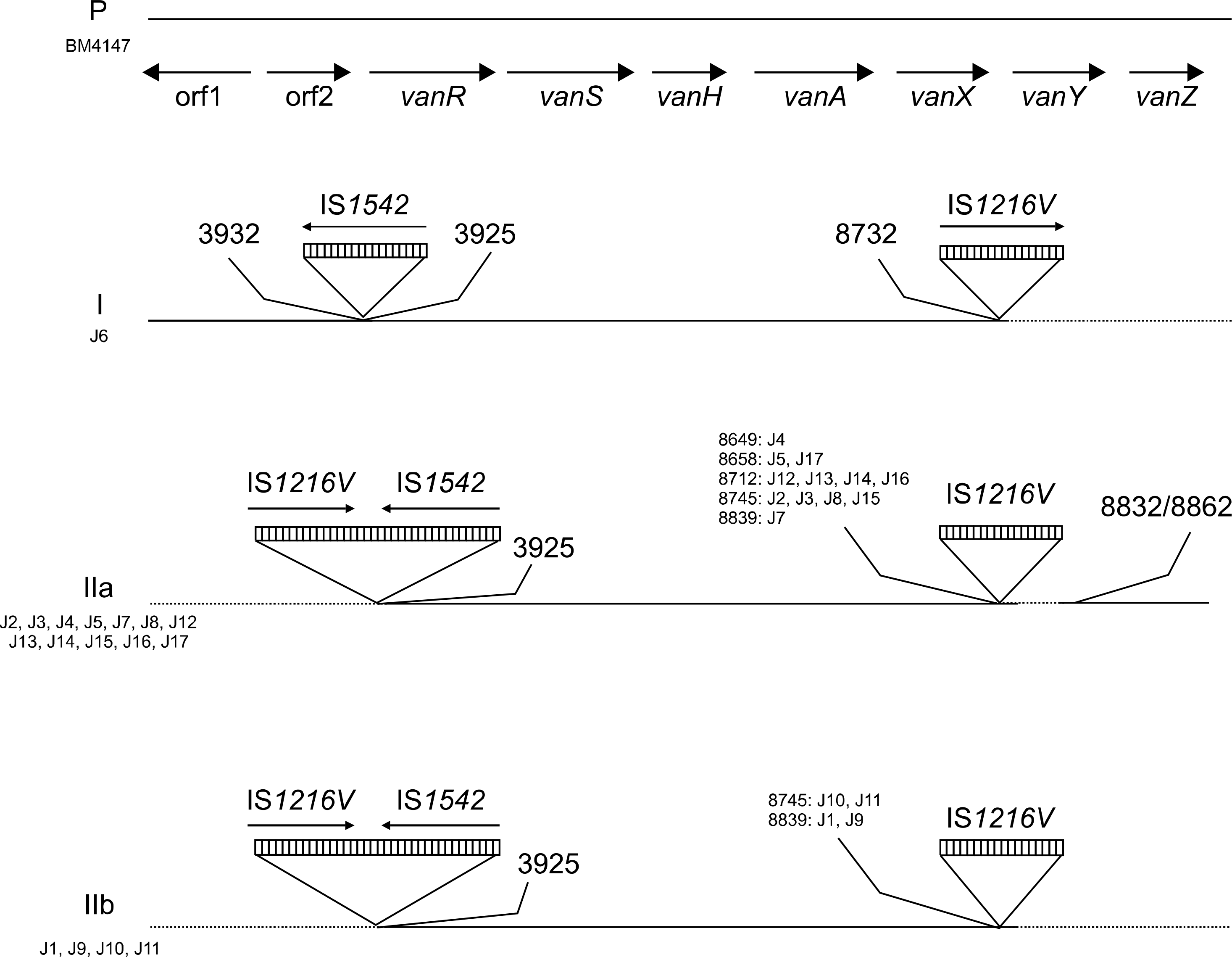 | Fig. 2.Tn1546 type of 17 vanA positive E. faecalis isolates. The positions of the genes and open reading frames (orf1 and orf2) and the direction of transcription are marked by big arrows at the top. Inverted triangles represent IS elements. The position of the first nucleotide upstream and the first nucleotide downstream from the IS insertion sites are depicted. Small arrows indicate the transcriptional orientation of the inserted IS elements. Deletions are indicated by dotted lines. Adapted from “Genetic rearrangements of Tn1546-like elements in vancomycin-resistant Enterococcus faecium isolates collected from hospitalized patients over a seven-year period,” by Park IJ, Lee WG, Lim YA and Cho SR, J Clin Microbiol 2007;45:3903-8. Adapted with permission. |
Table 1.
| Strain | Specimen | esp gen analysis | Tn 1546 type | PFGE based dendrogram | |||||
|---|---|---|---|---|---|---|---|---|---|
| 100% | 80% | 60% | 40% | 20% | 0% | ||||
| J1 | Catheter | Neg∗ | IIb | @@@image@@@@ | |||||
| J3 | Wound | Pos† | IIa | ||||||
| J12 | Wound | Pos† | IIa | ||||||
| J2 | Wound | Neg∗ | IIa | ||||||
| J4 | Urine | Pos† | IIa | ||||||
| J5 | Urine | Pos† | IIa | ||||||
| J8 | Blood | Neg∗ | IIa | ||||||
| J6 | Wound | Pos† | I | ||||||
| J9 | Urine | Neg∗ | IIb | ||||||
| J10 | Catheter | Neg∗ | IIb | ||||||
| J13 | Wound | Pos† | IIa | ||||||
| J17 | Urine | Neg∗ | IIa | ||||||
| J15 | Wound | Pos† | IIa | ||||||
| J14 | Pus | Neg∗ | IIa | ||||||
| J16 | Wound | Pos† | IIa | ||||||
| J7 | Urine | Pos† | IIa | ||||||
| J11 | Urine | Pos† | IIb | ||||||




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download