초록
Background
Fluoroquinolones (FQs) are important drugs for treating multidrug-resistant tuberculosis (MDR-TB). However, due to widespread use of FQs, the resistance rates to FQs have been increasing among Mycobacterium tuberculosis. Rapid and reliable FQ drug susceptibility testing (DST) is crucial for successful treatment of MDR-TB. In this study, the feasibility of molecular detection of FQ resistance was evaluated.
Methods
A total of 95 MDR-TB isolates were collected from Jan through Oct 2009 at the Korean Institute of Tuberculosis. DST for ofloxacin (OFL), levofloxacin, and moxifloxacin was performed using the Lowenstein-Jensen media absolute concentration method. Minimum inhibitory concentrations (MIC) of these were determined using the broth microdilution method. DNA was extracted from cultured isolates using bead beating method. The quinolone resistance-determining region (QRDR) of gyrA and gyrB were amplified and those sequences were analyzed.
Results
Of 95 isolates, 79 were resistant to at least one of FQs. Of these, 71 (89.9%) harbored mutation in the QRDR of gyrA or gyrB. None of FQ susceptible strains possessed any mutation in gyrA or gyrB. Mutations in codon 94 of gyrA were most common; only two isolates had mutation in only the gyrB gene. OFL MICs for isolates with gyrA mutation ranged from 1 to 32 μ g/mL, but FQ susceptible isolates showed MICs ranging from ≤0.06 to 0.5 μ g/mL.
Go to : 
REFERENCES
1.WHO. Global tuberculosis control: WHO report 2012. Geneva, Switzerland: WHO press. 2013.
2.Johnston JC., Shahidi NC., Sadatsafavi M., Fitzgerald JM. Treatment outcomes of multidrug-resistant tuberculosis: a systematic review and meta-analysis. PLoS One. 2009. 4:e6914.

3.Devasia RA., Blackman A., Gebretsadik T., Griffin M., Shintani A., May C, et al. Fluoroquinolone resistance in Mycobacterium tuberculosis: the effect of duration and timing of fluoroquinolone exposure. Am J Respir Crit Care Med. 2009. 180:365–70.
4.Kim SJ. Drug-susceptibility testing in tuberculosis: methods and reliability of results. Eur Respir J. 2005. 25:564–9.

5.Boehme CC., Nabeta P., Hillemann D., Nicol MP., Shenai S., Krapp F, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010. 363:1005–15.

6.Barnard M., Albert H., Coetzee G., O'Brien R., Bosman ME. Rapid molecular screening for multidrug-resistant tuberculosis in a high-volume public health laboratory in South Africa. Am J Respir Crit Care Med. 2008. 177:787–92.

7.van Ingen J., Simons S., de Zwaan R., van der Laan T., Kamst-van Agterveld M., Boeree MJ, et al. Comparative study on genotypic and phenotypic second-line drug resistance testing of Mycobacterium tuberculosis complex isolates. J Clin Microbiol. 2010. 48:2749–53.
8.Kiet VS., Lan NT., An DD., Dung NH., Hoa DV., van Vinh Chau N, et al. Evaluation of the MTBDR sl test for detection of second- line-drug resistance in Mycobacterium tuberculosis. J Clin Microbiol. 2010. 48:2934–9.
9.Hillemann D., Rüsch-Gerdes S., Richter E. Feasibility of the GenoType MTBDR sl assay for fluoroquinolone, amikacin-capre-omycin, and ethambutol resistance testing of Mycobacterium tuberculosis strains and clinical specimens. J Clin Microbiol. 2009. 47:1767–72.
10.Campbell PJ., Morlock GP., Sikes RD., Dalton TL., Metchock B., Starks AM, et al. Molecular detection of mutations associated with first- and second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2011. 55:2032–41.
11.Brossier F., Veziris N., Aubry A., Jarlier V., Sougakoff W. Detection by GenoType MTBDR sl test of complex mechanisms of resistance to second-line drugs and ethambutol in multidrug-resistant Mycobacterium tuberculosis complex isolates. J Clin Microbiol. 2010. 48:1683–9.
12.Sekiguchi J., Miyoshi-Akiyama T., Augustynowicz-Kopeć E., Zwolska Z., Kirikae F., Toyota E, et al. Detection of multidrug resistance in Mycobacterium tuberculosis. J Clin Microbiol. 2007. 45:179–92.
13.Wang JY., Lee LN., Lai HC., Wang SK., Jan IS., Yu CJ, et al. Fluoroquinolone resistance in Mycobacterium tuberculosis isolates: associated genetic mutations and relationship to antimicrobial exposure. J Antimicrob Chemother. 2007. 59:860–5.
14.CLSI. Susceptibility testing of mycobacteria, Nocardiae, and other aerobic actinomycetes; approved standard-second edition. CLSI document M24-A2. Wayne, PA: Clinical and Laboratory Standards Institute. 2011.
15.Yew WW., Lange C. Fluoroquinolone resistance in Mycobacterium tuberculosis. What have we learnt? Int J Tuberc Lung Dis. 2014. 18:1–2.
16.Kwon YS., Kim YH., Suh GY., Chung MP., Kim H., Kwon OJ, et al. Treatment outcomes for HIV-uninfected patients with multi-drug-resistant and extensively drug-resistant tuberculosis. Clin Infect Dis. 2008. 47:496–502.

17.Bai GH., Park YK., Choi YW., Bai JI., Kim HJ., Chang CL, et al. Trend of antituberculosis drug resistance in Korea, 1994-2004. Int J Tuberc Lung Dis. 2007. 11:571–6.
Go to : 
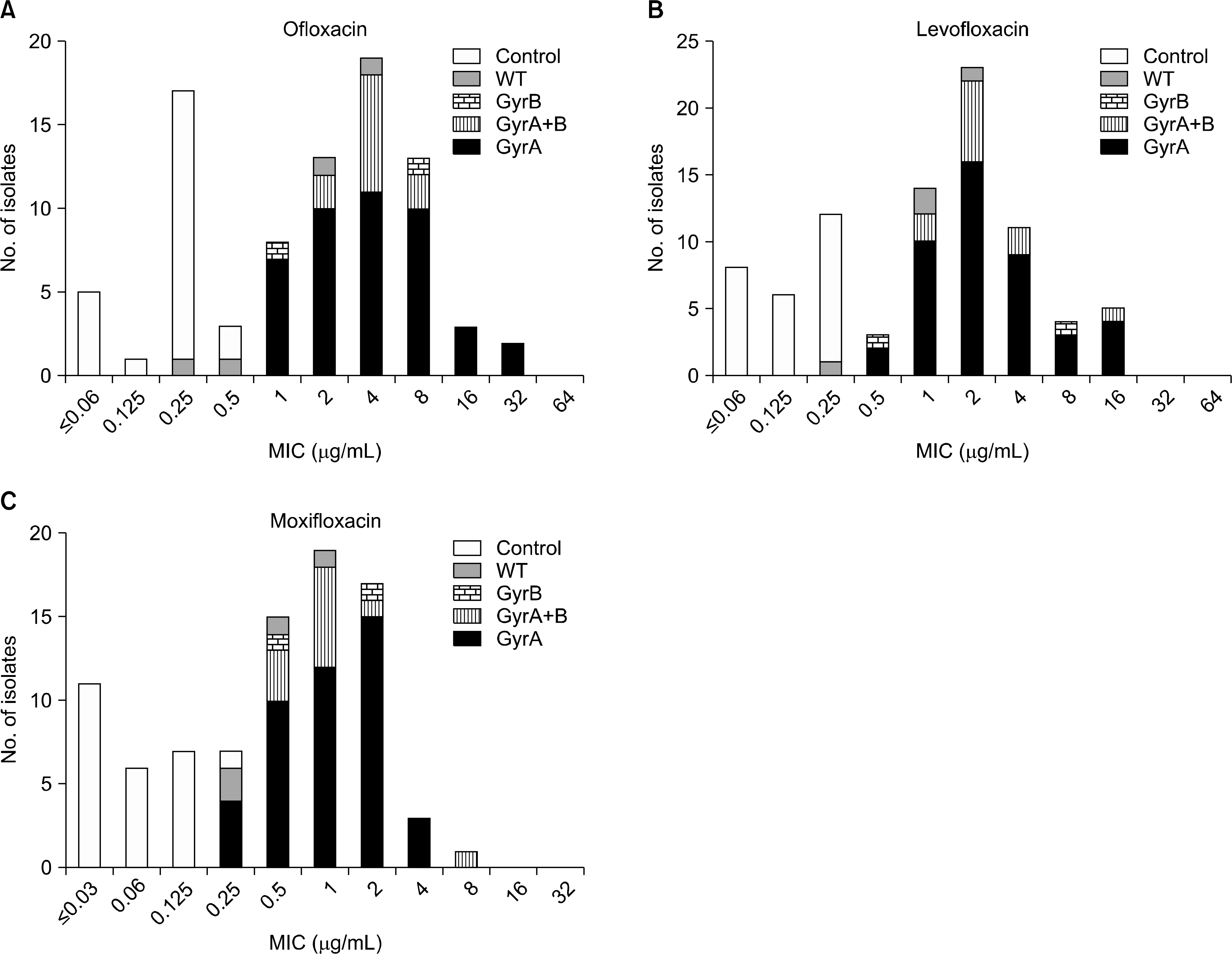 | Fig. 1.Distribution of fluoroquinolone minimum inhibitory concentration (MIC) of FQ-resistant multidrug-resistant tuberculosis (MDR-TB) isolates (n=71) and FQ-susceptible control isolates (n=25) according to gyrA and gyrB mutation patterns. This figure displays the number of isolates at each MIC and mutation patterns. The MICs of FQ-resistant isolates are close to but clearly discriminated from those of FQ-susceptible isolates. There is no apparent distinction in the MIC distribution between gyrA and gyrB mutation groups. (GyrA, GyrB and GyrA+B) (A: Ofloxacin, B: Levofloxacin, C: Moxifloxacin.) Abbreviations: SC, susceptible control; WT, wild type; gyrB, isolates with mutation in gyrB; gyrA, isolates with mutation in gyrB; gyrA+B , isolates with mutations in both gyrA and gyrB.
|
Table 1.
Primers used for PCR amplification and sequencing
Table 2.
Mutations in gyrA and gyrB genes of multidrug-resistant tuberculosis isolates and susceptible isolates




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download