초록
Background
Testing for possible microorganism contamination in umbilical cord blood (UCB) is essential for validating the product safety of allogeneic cellular therapeutics. We analyzed the level of contamination and related factors at the largest public cord blood bank in Korea. In addition, we also studied the influ-ence of cryopreservation on contaminating microorganisms.
Methods
UCB was collected, transported, processed, and stored according to standard operating procedures. Microbial detection and identification was performed using a conventional automated blood culture system (BacT/ALERT; bioMérieux, France) with an inoculum of 5-10 mL plasma for pre-freezing UCB. Forty ran-domly selected non-conforming units were thawed and studied for microbiologic recovery with an inoculum of 2.5 mL.
Results
Among a total of 21,236 UCB, 677 (3.19%) were positive for culture. The most frequently identified organism was Lactobacillus spp. (17.2%), followed by Bacteroides spp. (10.1%), coagulase negative staphylococcus (6.4%), except the unidentified gram positive bacillus (21.4%). The contamination rate was higher in vaginal delivery specimens than in cesar-ean section specimens (4.1% vs. 0.7%, P<0.001), and differed by collection center (0.7-25.4%, P<0.001). Only 55% after-thaw cultures of non-conforming units were positive.
Conclusion
We determined the contamination rate of UCB in Korea in a large sample size. The results of this study could be used as baseline data at collection centers for quality control purposes. The low recovery rate of microorganisms after cryopreservation presents a possible way to rescue some non-con-forming cord blood units, although further study is needed to confirm the reduction of microbiological burden.
Go to : 
REFERENCES
1.U.S. Department of Health and Human Services, Food and Drug Administration, Center for Biologics Evaluation and Research. Guidance for Industry: Minimally Manipulated, Unrelated Allogeneic Placental/Umbilical Cord Blood Intended for Hematopoietic Recon-stitution for Specified Indications. Rockville: Office of Communi-cation, Outreach and Development (OCOD);2009.
2.American Association of Blood Banks. Standards for Cellular Therapy Product Services. 5th ed. Bethesda Md.: AABB;2011.
3.Foundation for the Accreditation of Cellular Therapy (FACT). NetCord-FACT International Standards for Cord Blood Collection, Banking, and Release for Administration. 4th ed.http://www.factweb.org/[Online. (last visited on 7 August 2012).
4.Umbilical Cord Blood Management and Research Act. Korea Ministry of Health and Welfare, 2011. http://law.go.kr/[Online. (last visited on 7 August 2012).
5.Clark P., Trickett A., Stark D., Vowels M. Factors affecting microbial contamination rate of cord blood collected for transplantation. Transfusion. 2012. 52:1770–7.

6.Kamble R., Pant S., Selby GB., Kharfan-Dabaja MA., Sethi S., Kratochvil K, et al. Microbial contamination of hematopoietic progenitor cell grafts-incidence, clinical outcome, and cost-effecti-veness: an analysis of 735 grafts. Transfusion. 2005. 45:874–8.

7.Honohan A., Olthuis H., Bernards AT., van Beckhoven JM., Brand A. Microbial contamination of cord blood stem cells. Vox Sang. 2002. 82:32–8.

8.Morrow JF., Braine HG., Kickler TS., Ness PM., Dick JD., Fuller AK. Septic reactions to platelet transfusions. A persistent problem. JAMA. 1991. 266:555–8.

9.Tipple MA., Bland LA., Murphy JJ., Arduino MJ., Panlilio AL., Farmer JJ 3rd, et al. Sepsis associated with transfusion of red cells contaminated with Yersinia enterocolitica. Transfusion. 1990. 30:207–13.
10.Versalovic J, American Society for Microbiology. eds. Manual of Clinical Microbiology. 10th ed. Washington, DC: ASM Press;2011.
11.McDonald CP., Rogers A., Cox M., Smith R., Roy A., Robbins S, et al. Evaluation of the 3D BacT/ALERT automated culture system for the detection of microbial contamination of platelet concentrates. Transfus Med. 2002. 12:303–9.

12.Armitage S., Warwick R., Fehily D., Navarrete C., Contreras M. Cord blood banking in London: the first 1000 collections. Bone Marrow Transplant. 1999. 24:139–45.

13.Mermel LA., Maki DG. Detection of bacteremia in adults: consequences of culturing an inadequate volume of blood. Ann Intern Med. 1993. 119:270–2.

14.Basch H., Gadebusch HH. In vitro antimicrobial activity of dimethylsulfoxide. Appl Microbiol. 1968. 16:1953–4.

15.Kahn RA., Meryman HT., Syring RL., Flinton LJ. The fate of bacteria in frozen red cells. Transfusion. 1976. 16:215–20.

16.Roh EY., Yoon JH., Chang JY., Hwang KR., Song EY., Shin S. Analysis of mycoplasma contamination in donated cord blood units. Korean J Blood Transfus. 2008. 19:9–14.
17.Roh EY., Shin S., Yoon JH., Chang JY. Ureaplasma contamination rate in donated cord blood units. Korean J Blood Transfus. 2008. 19:239–44.
Go to : 
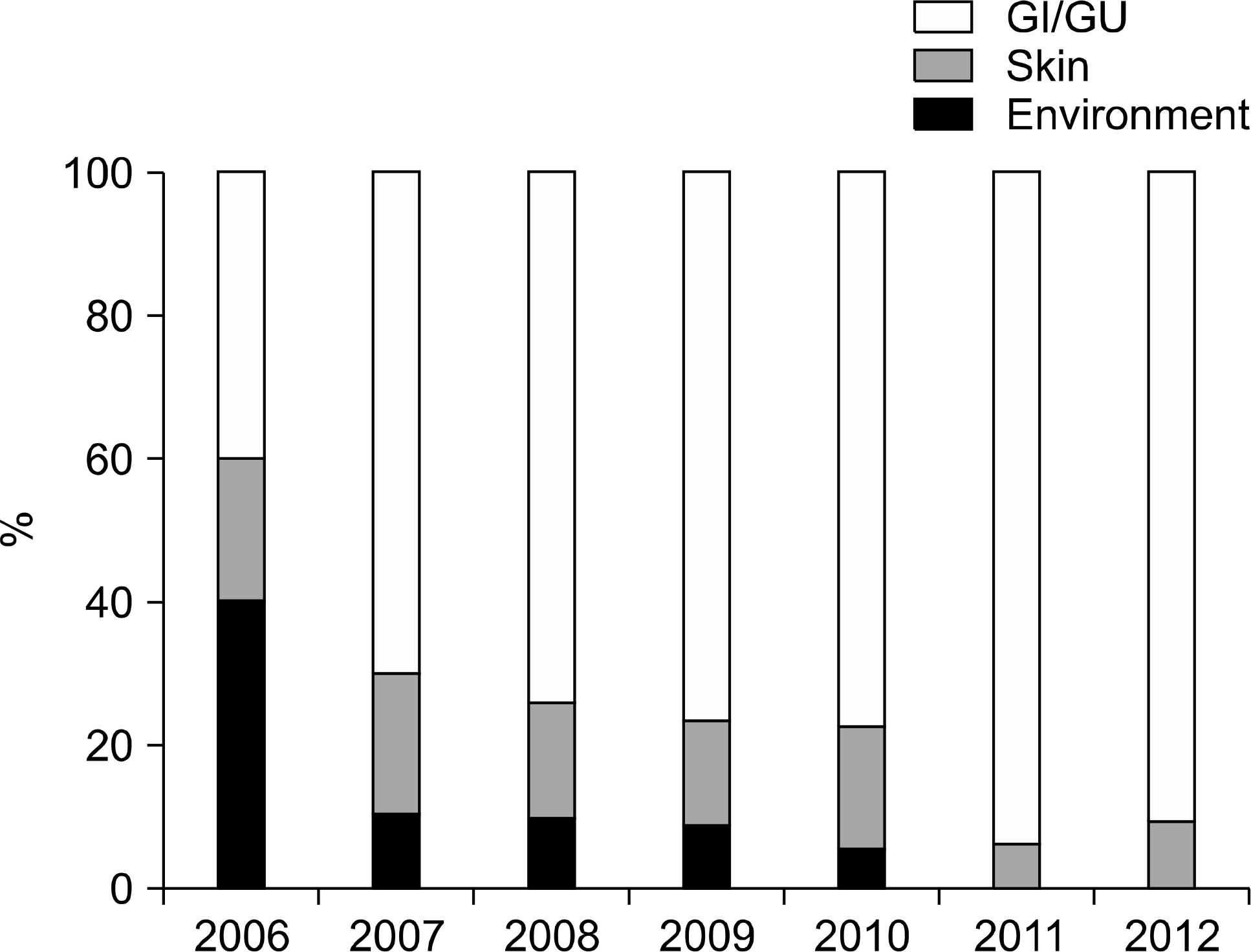 | Fig. 1.Proportions of origin of contaminated microorganisms according to years. Abbreviations: GI, gastrointestinal; GU, genitourinary. |
Table 1.
Micro-organisms isolated from culture positive umbilical cord blood units
| Potential contamination source | Frequency (%) |
|---|---|
| Skin flora | 11.0 |
| Coagulase negative staphylococcus | 6.3 |
| Corynebacterium spp. | 3.1 |
| Staphylococcus aureus | 0.7 |
| Propionibacterium acnes | 0.4 |
| Micrococcus spp. | 0.4 |
| Gastrointestinal/urogenital flora∗ | 55.8 |
| Lactobacillus spp. | 17.1 |
| Bacteroides spp. | 10.1 |
| Escherichia coli | 4.2 |
| Enterococcus spp. | 4.1 |
| Streptococcus viridans group | 3.9 |
| Peptostreptococcus spp. | 3.4 |
| Prevotella spp. | 3.4 |
| Lactococcus spp. | 2.1 |
| Klebsiella spp. | 1.9 |
| Clostridium spp. | 1.2 |
| Environmental contaminants∗ | 5.5 |
| Citrobacter spp. | 1.2 |
| Pseudomonas spp. | 1.0 |
| Unidentified Gram positive bacilli | 21.3 |
| Unidentified organisms | 4.0 |
| Missing data | 2.4 |
Table 2.
Contamination rate of umbilical cord blood unit according to collection center
| Collection center | Culture positive units (N) | Stored units (N) | Contamination rate (%) | Vaginal delivery rate (%) |
|---|---|---|---|---|
| A | 28 | 1,795 | 1.6 | 77.5 |
| B | 65 | 1,721 | 3.8 | 61.4 |
| C | 11 | 1,063 | 1.0 | 59.4 |
| D | 20 | 786 | 2.5 | 77.6 |
| E | 17 | 766 | 2.2 | 75.2 |
| F | 12 | 669 | 1.8 | 78.3 |
| G | 31 | 599 | 5.2 | 67.8 |
| H | 127 | 501 | 25.3∗ | 73.7 |
| I | 15 | 485 | 3.1 | 84.5 |
| J | 29 | 483 | 6.0 | 51.1 |
| K | 26 | 473 | 5.5 | 82.9 |
| L | 7 | 414 | 1.7 | 70.0 |
| M | 10 | 385 | 2.6 | 75.8 |
| N | 9 | 364 | 2.5 | 79.1 |
| O | 6 | 360 | 1.7 | 66.7 |
| P | 15 | 347 | 4.3 | 74.1 |
| Q | 20 | 320 | 6.3 | 70.3 |
| R | 2 | 309 | 0.6 | 74.8 |
Table 3.
Univariate analysis for parameters associated with the contamination of cord blood units
| Culture positive | Culture negative | P | |
|---|---|---|---|
| Type of delivery, N (%) | |||
| Vaginal delivery | 634 (4.1) | 14,987 (95.9) | <0.001 |
| Cesarean section | 37 (0.7) | 5,420 (99.3) | |
| Collection center, N (%) | |||
| H center∗ | 127 (25.3) | 374 (74.7) | <0.001 |
| Other centers | 550 (2.7) | 20,176 (97.3) | |
| Maternal age (years)† | 31 (29-34) | 31 (29-33) | 0.728 |
| Gestational age (months)† | 39 (39-40) | 39 (38-40) | 0.067 |
| Birth weight (kg)† | 3.35 (3.12-3.60) | 3.34 (3.10-3.60) | 0.852 |
| Volume of cord blood (mL)† | 105.8 (95.2-118.9) | 108.0 (96.6-122.0) | <0.001 |
| Processing time (hours)† | 25 (22-29) | 26 (23-30) | 0.003 |
| White blood cell (×103/uL)† | 10.7 (9.3-12.9) | 10.6 (8.9-12.6) | 0.006 |
| Platelet (×103/uL)† | 210 (186-238) | 208 (183-234) | 0.089 |
| Hemoglobin (g/dL)† | 11.6 (10.8-12.4) | 11.7 (10.9-12.6) | 0.125 |
| Nucleated RBC (/100 WBC)† | 2.2 (1.2-4.7) | 2.0 (1.0-3.9) | <0.001 |
Table 4.
Multivariate logistic regression for parameters associated with the contamination of cord blood units
| Beta coefficient | Odds ratio | 95% Confidence interval | P | |
|---|---|---|---|---|
| Vaginal delivery∗ | 1.938 | 6.942 | 4.891-9.854 | <0.001 |
| H center† | 2.548 | 12.777 | 10.160-16.068 | <0.001 |
| Volume of cord blood (mL) | -0.007 | 0.993 | 0.988-0.998 | 0.003 |
| Processing time (hours) | -0.011 | 0.989 | 0.979-1.000 | 0.048 |
| Hemoglobin (g/dL) | -0.06 | 0.942 | 0.882-1.005 | 0.069 |
| Nucleated RBC (/100 WBC) | 0.02 | 1.02 | 1.006-1.033 | 0.004 |
Table 5.
Discordant culture results of contaminated cord blood units between pre-freeze and post-thaw




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download