초록
Background
Cytomegalovirus (CMV) infection is a major cause of morbidity and mortality in immunoco-mpromised patients. We compared the abilities of the recently developed Real-Q Cytomegalovirus Kit (Bio-sewoom Inc., Korea) and the previously used PANA mPCR CMV Detection Kit (Panagene Inc., Korea) to detect CMV.
Methods
We analyzed 300 samples (whole blood: 262, urine: 37, CSF: 1) submitted for qualitative CMV PCR testing during October 2011 at Yonsei University College of Medicine Severance Hospital. real-time PCR was performed with a Real-Q Cytomegalovirus Kit and conventional PCR was conducted with a PANAmPCR CMV Detection Kit.
Results
The positive rates of both real-time PCR and conventional PCR were 25.3% (76/300), and the kappa coefficient (K) was 0.96 (95% confidence interval (CI), 0.93-1.00). The concordance rate of the Real-Q Cytomegalovirus Kit and the PANAmPCR CMV Detection Kit was 98.7% (296/300), and four out of 300 samples showed discordant results. If the concordant results of 296 samples and the four results confirmed by direct sequencing were assumed to be true, the sensitivity and specificity of the Real-Q Cytomegalovirus Kit were 97.4% (95% CI, 93.8-100.0%) and 99.1% (95% CI, 97.9-100.0%), respectively.
Conclusion
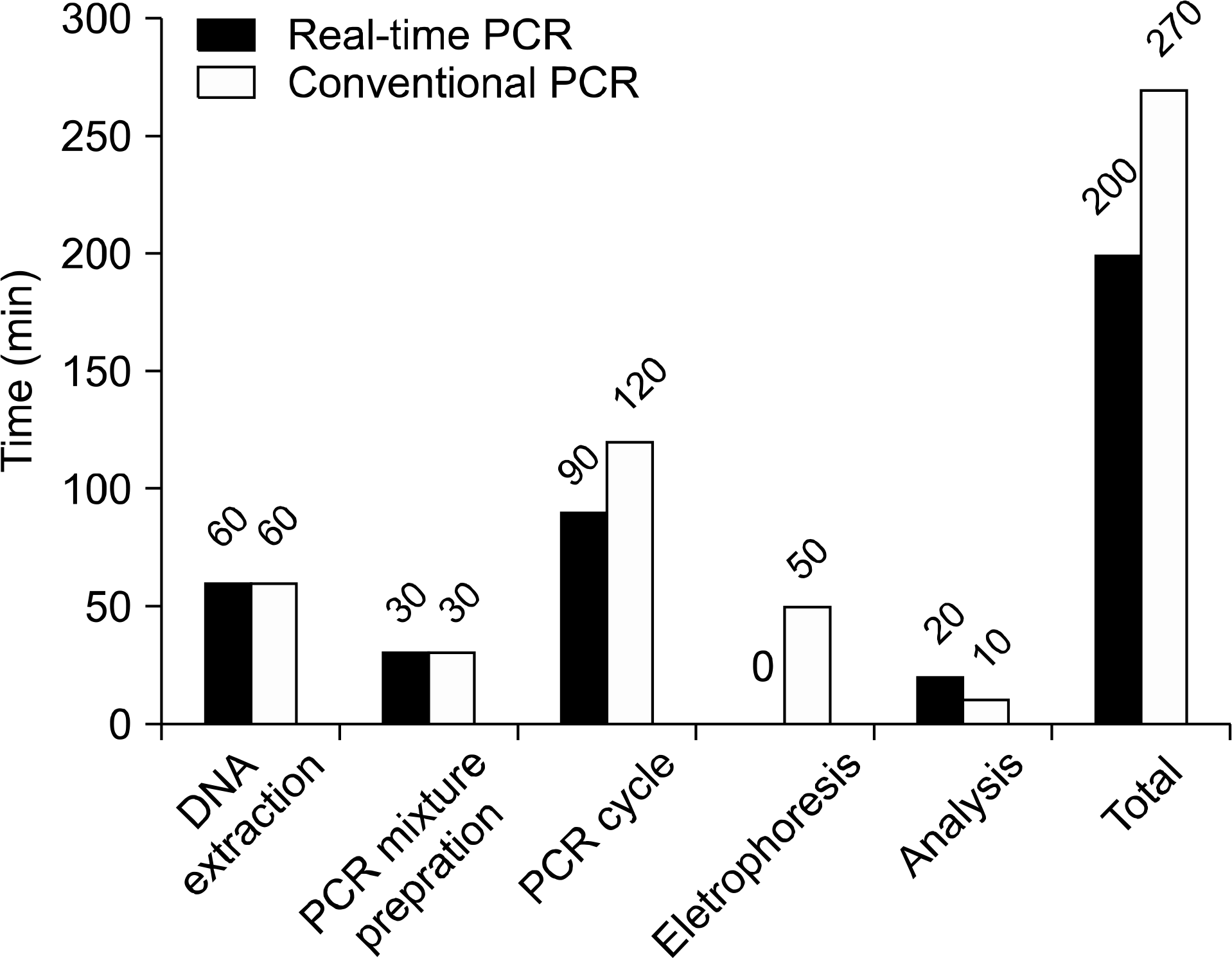
The recently developed Real-Q Cytome-galovirus Kit showed excellent sensitivity and specificity, and had a high concordance rate with the previously established PANAmPCR CMV Detection Kit, which uses conventional PCR. Furthermore, real-time PCR could decrease the test time, as the electro-phoresis step required for conventional PCR is not required for real-time PCR.
REFERENCES
1.Paya CV. Prevention of cytomegalovirus disease in recipients of solid-organ transplants. Clin Infect Dis. 2001. 32:596–603.
2.van Son WJ., The TH. Cytomegalovirus infection after organ transplantation: an update with special emphasis on renal transplantation. Transpl Int. 1989. 2:147–64.
3.Jung KW., Park S., Kong HJ., Won YJ., Boo YK., Shin HR, et al. Cancer statistics in Korea: incidence, mortality and survival in 2006-2007. J Korean Med Sci. 2010. 25:1113–21.

4.Cho WH., Kim SI., Kim MS., Ahn C., Bang KT., Jeon KO, et al. A proposal to activate organ donation: report of organ allocation study group. J Korean Soc Transplant. 2009. 23:8–14.
5.Lautenschlager I., Suni J., Ahonen J., Grönhagen-Riska C., Ruutu P., Ruutu T, et al. Detection of cytomegalovirus by the early-antigen immunofluorescence test versus conventional tissue culture. Eur J Clin Microbiol Infect Dis. 1989. 8:610–3.

6.Sheehan MM., Coker R., Coleman DV. Detection of cytomegalovirus (CMV) in HIV+ patients: comparison of cytomorphology, immu-nocytochemistry and in situ hybridization. Cytopathology. 1998. 9:29–37.
7.Boeckh M., Boivin G. Quantitation of cytomegalovirus: me-thodologic aspects and clinical applications. Clin Microbiol Rev. 1998. 11:533–54.

8.Lee YW., Nam MH., Lee JH., Lee NY. Comparison of polymerase chain reaction method and CMV antigenemia assay for diagnosis of cytomegalovirus infection in transplanted patients. Korean J Clin Microbiol. 1999. 2:177–81.
9.Guiver M., Fox AJ., Mutton K., Mogulkoc N., Egan J. Evaluation of CMV viral load using TaqMan CMV quantitative PCR and comparison with CMV antigenemia in heart and lung transplant recipients. Transplantation. 2001. 71:1609–15.

10.Delgado R., Lumbreras C., Alba C., Pedraza MA., Otero JR., Gómez R, et al. Low predictive value of polymerase chain reaction for diagnosis of cytomegalovirus disease in liver transplant recipients. J Clin Microbiol. 1992. 30:1876–8.

11.Kearns AM., Guiver M., James V., King J. Development and evaluation of a real-time quantitative PCR for the detection of human cytomegalovirus. J Virol Methods. 2001. 95:121–31.

12.Allice T., Enrietto M., Pittaluga F., Varetto S., Franchello A., Mar-chiaro G, et al. Quantitation of cytomegalovirus DNA by real-time polymerase chain reaction in peripheral blood specimens of patients with solid organ transplants: comparison with end-point PCR and pp65 antigen test. J Med Virol. 2006. 78:915–22.

13.Lisboa LF., Preiksaitis JK., Humar A., Kumar D. Clinical utility of molecular surveillance for cytomegalovirus after antiviral prophy-laxis in high-risk solid organ transplant recipients. Transplantation. 2011. 92:1063–8.

14.Seo S., Cho Y., Park J. Serologic screening of pregnant Korean women for primary human cytomegalovirus infection using IgG avidity test. Korean J Lab Med. 2009. 29:557–62.

15.Kim YK., Kim DW. Seroepidemiology of human cytomegalovirus in healthy adults measured by means of the anticomplement immunofluorescence technique. Korean J Infect Dis. 1992. 24:87–92.
16.Tun N., London N., Kyaw MK., Smithuis F., Ford N., Margolis T, et al. CMV retinitis screening and treatment in a resource-poor setting: three-year experience from a primary care HIV/AIDS programme in Myanmar. J Int AIDS Soc. 2011. 14:41.

17.Schmidt U., Metz KA., Soukou C., Quabeck K. The association of pulmonary CMV infection with interstitial pneumonia after bone marrow transplantation. Histopathological and immunohistochemical findings in 104 autopsies. Zentralbl Pathol. 1993. 139:225–30.
18.Engelhard D., Naparstek E., Or R., Nagler A., Jacobs J., Shahar MB, et al. Ganciclovir for the treatment of disseminated CMV disease without pneumonia in allogeneic T-lymphocyte depleted bone marrow transplantation. Leuk Lymphoma. 1993. 10:143–6.

19.Shin HJ., Kim HK., Kim HS. The usefulness of PCR and early antigen immunostaining as a rapid identification method of cytomegalovirus infection. Korean J Clin Pathol. 1998. 18:452–7.
20.Choe WH., Kang JO., Choi TY., Ahn Y. Detection of cytomegalovirus by Dual-PCR. Korean J Clin Microbiol. 2006. 9:96–101.
21.Li H., Dummer JS., Estes WR., Meng S., Wright PF., Tang YW. Measurement of human cytomegalovirus loads by quantitative real- time PCR for monitoring clinical intervention in transplant recipients. J Clin Microbiol. 2003. 41:187–91.
22.Boeckh M., Huang M., Ferrenberg J., Stevens-Ayers T., Stensland L., Nichols WG, et al. Optimization of quantitative detection of cytomegalovirus DNA in plasma by real-time PCR. J Clin Microbiol. 2004. 42:1142–8.

23.Miller MJ., Bovey S., Pado K., Bruckner DA., Wagar EA. Appli-cation of PCR to multiple specimen types for diagnosis of cytomegalovirus infection: comparison with cell culture and shell vial assay. J Clin Microbiol. 1994. 32:5–10.

24.Beutler E., Gelbart T., Kuhl W. Interference of heparin with the polymerase chain reaction. Biotechniques. 1990. 9:166.
25.Gerna G., Zipeto D., Parea M., Revello MG., Silini E., Percivalle E, et al. Monitoring of human cytomegalovirus infections and ganci-clovir treatment in heart transplant recipients by determination of viremia, antigenemia, and DNAemia. J Infect Dis. 1991. 164:488–98.

26.Drouet E., Colimon R., Michelson S., Fourcade N., Niveleau A., Ducerf C, et al. Monitoring levels of human cytomegalovirus DNA in blood after liver transplantation. J Clin Microbiol. 1995. 33:389–94.

Table 1.
Demographic characteristics of the patients
| Characteristics | No. of patients | Percentage (%) |
|---|---|---|
| Sex | ||
| Male | 168 | 56.0 |
| Female | 132 | 44.0 |
| Age (years) | ||
| Under 20 | 48 | 16.0 |
| 20-30 | 26 | 8.7 |
| 30-40 | 42 | 14.0 |
| 40-50 | 40 | 13.3 |
| 50-60 | 73 | 24.3 |
| 60 and over | 71 | 23.7 |
| Ward | ||
| ICU | 37 | 12.3 |
| General | 263 | 87.7 |
| Diagnosis | ||
| Hematologic disease | 175 | 58.3 |
| Solid cancer | 13 | 4.3 |
| Renal disease | 23 | 7.7 |
| Other∗ | 89 | 29.7 |
| Specimen type | ||
| Whold blood | 262 | 87.3 |
| Urine | 37 | 12.3 |
| CSF | 1 | 0.3 |
Table 2.
Comparison of real-time PCR and conventional PCR by kappa agreement and McNemar test for detection of CMV infection
| Parameters | Real-time PCR (n=300) | ||
|---|---|---|---|
| Positive | Negative | ||
| Conventional PCR | Positive | 74 | 2 |
| Negative | 2 | 222 | |
| Kappa agreement∗ | 0.96 (0.93 to 1.00) | ||
| P value of McNemar test | 1.0000 | ||
| Agreement (%) | 98.7 | ||
Table 3.
Results of CMV antibodies on the discrepancy (n=4) between real-time PCR and conventional PCR
| Sample No. | Real-time | Ct of real-time | Conventional | CMV IgM | CMV IgG | Direct sequencing∗ | ∗ Diagnosis | CMV associated diagnosis∗ |
|---|---|---|---|---|---|---|---|---|
| 1 | Negative | -† | Positive | Negative | Positive | Positive | Hepatocellular carcinoma | Leukopenia, thrombocytopenia |
| 2 | Positive | 39.87 | Negative | Negative | Positive | -‡ | Chronic myelocytic leukemia | Leukopenia, thrombocytopenia, enteritis |
| 3 | Negative | -† | Positive | Negative | Positive | Positive | Acute lymphocytic leukemia | Leukopenia, thrombocytopenia, retinitis |
| 4 | Positive | 39.91 | Negative | Not tested | Not tested | -‡ | Hodgkin's disease | Leukopenia, thrombocytopenia |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download