초록
Background
Multidrug resistant (MDR) Acinetobacter baumannii has emerged as a significant infectious agent in hospitals worldwide. The purpose of this study was to determine the molecular characterization of MDR A. baumannii clinical isolates.
Methods
Two hundred eighty-five strains of non-du-plicated A. baumannii collected from March to Novem-ber 2011 from a university hospital laboratory located in the Wonju area of the Gangwon province of Korea were analyzed for MDR genes.
Results
All of the 285 imipenem-resistant A. baumannii isolates were encoded by a blaOXA-23-like gene, and all isolates with the blaOXA-23-like gene had the upstream element IS Aba1. The 16S rRNA methylase gene arm A was detected in 153 (50.2%) clinical isolates, but rmt A, rmt B, rmt C, rmt D and npm A were not detected in any isolates in the present study. The gene encoding aac(6')-Ib was the most prevalent aminoglycoside-modifying enzyme. The sequencing data for the quinolone resistance-determining region of gyrA and parC revealed the presence of Ser (TCA) 83 to Leu (TTA) and Ser (TCG) 80 to Leu (TTG) substitutions. All but one of the 285 A. baumannii isolates showed similar band patterns on re-petitive extragenic palindromic-PCR profiles.
Go to : 
REFERENCES
1.Lockhart SR., Abramson MA., Beekmann SE., Gallagher G., Riedel S., Diekema DJ, et al. Antimicrobial resistance among Gram- negative bacilli causing infections in intensive care unit patients in the United States between 1993 and 2004. J Clin Microbiol. 2007. 45:3352–9.
2.Higgins PG., Poirel L., Lehmann M., Nordmann P., Seifert H. OXA-143, a novel carbapenem-hydrolyzing class D beta-lactamase in Acinetobacter baumannii. Antimicrob Agents Chemother. 2009. 53:5035–8.
3.Kim CK., Lee Y., Lee H., Woo GJ., Song W., Kim MN, et al. Prevalence and diversity of carbapenemases among imipenem- nonsusceptible Acinetobacter isolates in Korea: emergence of a novel OXA-182. Diagn Microbiol Infect Dis. 2010. 68:432–8.
4.Bou G., Oliver A., Martínez-Beltrán J. OXA-24, a novel class D beta-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob Agents Chemother. 2000. 44:1556–61.
5.Afzal-Shah M., Woodford N., Livermore DM. Characterization of OXA-25, OXA-26, and OXA-27, molecular class D beta-lactamases associated with carbapenem resistance in clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 2001. 45:583–8.
6.Héritier C., Poirel L., Aubert D., Nordmann P. Genetic and functio-nal analysis of the chromosome-encoded carbapenem-hydrolyzing oxacillinase OXA-40 of Acinetobacter baumannii. Antimicrob Agents Chemother. 2003. 47:268–73.
7.Jeong SH., Bae IK., Park KO., An YJ., Sohn SG., Jang SJ, et al. Outbreaks of imipenem-resistant Acinetobacter baumannii producing carbapenemases in Korea. J Microbiol. 2006. 44:423–31.
8.Roh KH., Kim CK., Yum JH., Yong D., Jeong SH., Lim CS, et al. Carbapenem resistance mechanisms and molecular epidemiology of Acinetobacter spp. from four hospitals in Seoul and Gyeonggi province in 2006. Korean J Clin Microbiol. 2010. 13:27–33.
9.Segal H., Jacobson RK., Garny S., Bamford CM., Elisha BG. Exten-ded-10 promoter in IS Aba-1 upstream of bla OXA-23 from Acinetobacter baumannii. Antimicrob Agents Chemother. 2007. 51:3040–1.
10.Peleg AY., Seifert H., Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008. 21:538–82.
11.Turton JF., Ward ME., Woodford N., Kaufmann ME., Pike R., Livermore DM, et al. The role of IS Aba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol Lett. 2006. 258:72–7.
12.Bae IK., Jeong SH., Lee K. Carbapenem-Resistant Acinetobacter baumannii. Korean J Clin Microbiol. 2012. 15:1–8.
13.Yong D., Shin JH., Kim S., Lim Y., Yum JH., Lee K, et al. High prevalence of PER-1 extended-spectrum beta-lactamase-producing Acinetobacter spp. in Korea. Antimicrob Agents Chemother. 2003. 47:1749–51.
14.Kim JW., Heo ST., Jin JS., Choi CH., Lee YC., Jeong YG, et al. Characterization of Acinetobacter baumannii carrying bla OXA-23, bla PER-1 and arm A in a Korean hospital. Clin Microbiol Infect. 2008. 14:716–8.
15.Lee G., Lee JH., Lim K., Suh IB., Ryu SW., Eom YB, et al. Prevalence of multidrug-resistant Acinetobacter baumannii producing OXA-23-like from a healthcare facility of Gangwon Province, South Korea. Int J Antimicrob Agents. 2012. 39:452–3.
16.Vila J., Ruiz J., Goñi P., Marcos A., Jimenez de Anta T. Mutation in the gyr A gene of quinolone-resistant clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 1995. 39:1201–3.
17.Vila J., Ruiz J., Goñi P., Jimenez de Anta T. Quinolone-resistance mutations in the topoisomerase IV par C gene of Acinetobacter baumannii. J Antimicrob Chemother. 1997. 39:757–62.
18.Wisplinghoff H., Decker M., Haefs C., Krut O., Plum G., Seifert H. Mutations in gyr A and par C associated with resistance to fluoro-quinolones in epidemiologically defined clinical strains of Acinetobacter baumannii. J Antimicrob Chemother. 2003. 51:177–80.
19.Hamouda A., Amyes SG. Novel gyr A and par C point mutations in two strains of Acinetobacter baumannii resistant to ciprofloxacin. J Antimicrob Chemother. 2004. 54:695–6.
20.Vakulenko SB., Mobashery S. Versatility of aminoglycosides and prospects for their future. Clin Microbiol Rev. 2003. 16:430–50.

21.Fritsche TR., Castanheira M., Miller GH., Jones RN., Armstrong ES. Detection of methyltransferases conferring high-level resistance to aminoglycosides in enterobacteriaceae from Europe, North America, and Latin America. Antimicrob Agents Chemother. 2008. 52:1843–5.

22.Galimand M., Courvalin P., Lambert T. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob Agents Chemother. 2003. 47:2565–71.
23.Lee H., Yong D., Yum JH., Roh KH., Lee K., Yamane K, et al. Dissemination of 16S rRNA methylase-mediated highly amikacin- resistant isolates of Klebsiella pneumoniae and Acinetobacter baumannii in Korea. Diagn Microbiol Infect Dis. 2006. 56:305–12.
24.Dijkshoorn L., Van Harsselaar B., Tjernberg I., Bouvet PJ., Vanee-choutte M. Evaluation of amplified ribosomal DNA restriction analysis for identification of Acinetobacter genomic species. Syst Appl Microbiol. 1998. 21:33–9.
25.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-first Informational Supplement. Document M100-S21. Wayne, PA; Clinical and Laboratory Standards Institute. 2011.
26.Lee K., Lim YS., Yong D., Yum JH., Chong Y. Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-beta-lactamase-producing isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol. 2003. 41:4623–9.
27.Woodford N., Ellington MJ., Coelho JM., Turton JF., Ward ME., Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006. 27:351–3.
28.Segal H., Garny S., Elisha BG. Is IS ABA-1 customized for Acinetobacter? FEMS Microbiol Lett. 2005. 243:425–9.
29.Hujer KM., Hujer AM., Endimiani A., Thomson JM., Adams MD., Goglin K, et al. Rapid determination of quinolone resistance in Acinetobacter spp. J Clin Microbiol. 2009. 47:1436–42.
30.Akers KS., Chaney C., Barsoumian A., Beckius M., Zera W., Yu X, et al. Aminoglycoside resistance and susceptibility testing errors in Acinetobacter baumannii-calcoaceticus complex. J Clin Microbiol. 2010. 48:1132–8.
31.Lee K., Kim MN., Kim JS., Hong HL., Kang JO., Shin JH, et al. KONSAR Group. Further increases in carbapenem-, amikacin-, and fluoroquinolone-resistant isolates of Acinetobacter spp. and P. aeruginosa in Korea: KONSAR study 2009. Yonsei Med J. 2011. 52:793–802.
32.Lee K., Kim MN., Choi TY., Cho SE., Lee S., Whang DH, et al. KONSAR Group. Wide dissemination of OXA-type carbapene-mases in clinical Acinetobacter spp. isolates from South Korea. Int J Antimicrob Agents. 2009. 33:520–4.
33.Park YS., Lee H., Lee KS., Hwang SS., Cho YK., Kim HY, et al. Extensively drug-resistant Acinetobacter baumannii: risk factors for acquisition and prevalent OXA-type carbapenemases--a multicentre study. Int J Antimicrob Agents. 2010. 36:430–5.
34.Mendes RE., Bell JM., Turnidge JD., Castanheira M., Jones RN. Emergence and widespread dissemination of OXA-23, -24/40 and -58 carbapenemases among Acinetobacter spp. in Asia-Pacific nations: report from the SENTRY Surveillance Program. J Antimicrob Chemother. 2009. 63:55–9.
35.Koo SH., Kwon KC., Cho HH., Sung JY. Genetic basis of multidrug- resistant Acinetobacter baumannii clinical isolates from three university hospitals in Chungcheong Province, Korea. Korean J Lab Med. 2010. 30:498–506.
36.Cho YJ., Moon DC., Jin JS., Choi CH., Lee YC., Lee JC. Genetic basis of resistance to aminoglycosides in Acinetobacter spp. and spread of arm A in Acinetobacter baumannii sequence group 1 in Korean hospitals. Diagn Microbiol Infect Dis. 2009. 64:185–90.
Go to : 
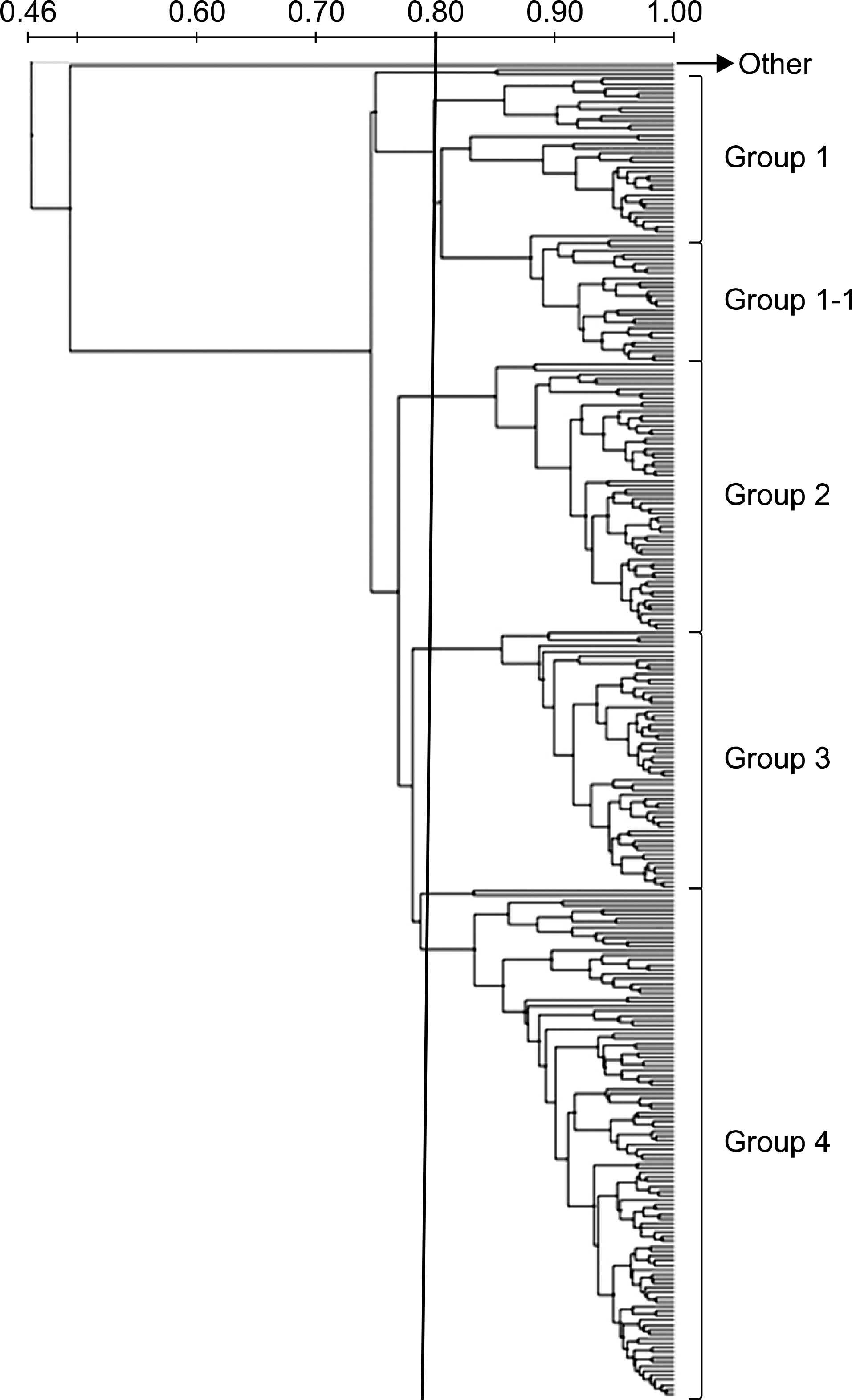 | Fig. 1.Dendrogram of the similarity index by unweighted pair group method with mathematical averaging (UPGMA) among two hundred sixty-five A. baumannii isolates provided by REP-PCR analysis. |
Table 1.
Antimicrobial resistance for 285 imipenem-resistant A. baumannii isolates
Table 2.
Distribution of quinolone resistant genotypes in A. baumannii
Table 3.
Distribution of aminoglycoside resistance genes in A. baumannii
Table 4.
Molecular characterization group of MDR A. baumannii by REP-PCR similarity index




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download