Abstract
Purpose
The specific aim of this study is to unravel a DNA copy number alterations, and to search for novel genes that are associated with the development of Korean gastric cancer.
Materials and Methods
We investigated a DNA copy number changes in 23 gastric adenocarcinomas by array-comparative genomic hybridization and quantitative real-time polymerase chain reaction analyses. Besides, the expression of UQCRFS1, which shows amplification in array-CGH, was examined in 186 gastric cancer tissues by an immunohistochemistry, and in 9 gastric cancer cell lines, as well as 24 gastric cancer tissues by immunoblotting.
Results
We found common gains at 48 different loci, and a common loss at 19 different loci. Amplification of UQCRFS1 gene at 19q12 was found in 5 (21.7%) of the 23 gastric cancers in an array-comparative genomic hybridization and DNA copy number were increased in 5 (20.0%) out of the 25 gastric cancer in quantitative real-time polymerase chain reaction. In immunohistochemistry, the overexpression of the protein was detected in 105 (56.5%) out of the 186 gastric cancer tissues. Statistically, there was no significant relationship between the overexpression of UQCRFS1 and clinicopathologic parameters (P>0.05). In parallel, the overexpression of UQCRFS1 protein was confirmed in 6 (66.7%) of the 9 gastric cancer cell lines, and 12 (50.0%) of the 24 gastric cancer tissues by immunoblotting.
Gastric cancer occurs with a high incidence in Asia and is one of the leading causes of cancer deaths worldwide.(1) In 2008, it accounts for an 15.7% of all cancer occurrences in Korea.(2) Therefore, gastric cancer remains a significant health burden worldwide. Surgical removal is the treatment of choice in advanced gastric cancer cases, whereas endoscopic mucosal resection can be curative in early gastric cancer. Detection and removal of gastric cancer in an early stage will contribute to reduce death due to gastric cancer. The clinical outcome of gastric cancer has gradually improved, but the prognosis of patients at the advanced stage is still poor. Thus, better understanding of the biology and development of molecular test for early detection of gastric cancer are crucial in this respect.
Many solid tumors, including gastric cancer, show defects in the maintenance of genomic stability, frequently resulting in DNA copy number alterations.(3,4) Comparative genomic hybridization (CGH) with total genomic DNA from test and reference cell population was the first efficient approach to scanning the entire genome for variations in DNA copy number. However, because of the difficulty in obtaining sufficient metaphase chromosome spreads for classical banding analyses from primary cancer tissues, there is little information on cytogenetic change of gastric cancer. ArrayCGH with elements produced by spotting DNA made from genomic clones, cDNAs, selected polymerase chain reaction (PCR) products, and oligonucleotides is a more powerful tool for the rapid and accurate detection of specific gene copy number change, allowing the identification of novel candidate genes in tumorigenesis. To date, a number of reports have described the CGH analysis of gastric cancers.(4-6) Interestingly, it has been reported that gastric cancer is a heterogenous disease at the genomic level and that different patterns of DNA copy number aberrations are correlated to differences in clinical behavior and patient outcome.(5,6) For example, the degree of genetic change at the DNA ploidy level correlated well with tumor stage in diffuse-type gastric cancers, while such a correlation was poor in intestinal-type gastric cancer.(7,8)
The present study aimed to unravel DNA copy number alterations in gastric cancers and to identify novel candidate genes that may contribute to the development or progression of gastric cancer. Therefore, we applied arrayCGH analysis to 23 gastric adenocarcinomas and found the UQCRFS1 candidate gene with high-level gains (amplification) and overexpression in gastric cancers.
Methacarn fixed paraffin-embedded, twenty three advanced gastric cancer specimens were enrolled in this study. Of them, 14 cases were of the intestinal-type, 5 were of the diffuse-type and 4 were of the mixed-type. Approval was obtained from the institutional review board of The Catholic University of Korea, College of Medicine (CUMC09U043). There was no evidence of familial cancer in any of the patients.
The tumor cells were selectively procured from Hematoxylin & Eosin stained slides using a laser microdissection device (ION LMD, JungWoo International Co, Seoul, Korea). The surrounding non-cancerous gastric mucosal cells were also obtained to study the corresponding normal DNA from the same slides in all cases. DNA extraction was performed using commercially available DNA isolation kits (AIAamp DNA mini kit; Qiagen, Hilden, Germany).
We used oligo arrayCGH microarrays (Agilent Technologies, Palo Alto, CA, USA), which contained over 43,000 probes representing coding and noncoding human sequences. Genomic DNAs extracted from the gastric cancers and corresponding gastric mucosae were labeled with Cy5 and Cy3, respectively, and competitively hybridized on the microarrays for 24 hours. After washing microarrays, the microarrays were scanned using GenePix 4000B microarray scanner (Molecular Devices, Sunnyvale, CA, USA) and Feature Extraction Software (Agilent Technologies) was used to calculate signal intensities of fluorescent dyes on each probes. Copy number alterations spanning less than three probes were excluded to increase specificity.
Primers were designed using the Primer3.(9) The amplification was performed in 20 µl containing 10 µl iQ SYBR Green Supermix (BioRad, Hercules, CA, USA), 10 µM of each primer and 1 µl of genomic DNA extracted from 25 frozen gastric cancer tissues. The PCR reactions were run in LightCycler 480 II (Roche Molecular Biochemicals, Mannheim, Germany) using the following program: 95℃ for 5 minutes and 45 cycles of 95℃ for 30 seconds and 60℃ for 30 seconds and 72℃ for 30 seconds. Two technical replicates of each reaction were performed and β-actin was used as internal control for normalization. The following oligonucleotide primer pairs were used: for UQCRFS1 5'-GGAAATTGAGCAGGAAGCTG-3' and 5'-GGCAAGGGCAGTAATAACCA-3'; for β-actin 5'-GATGAGATTGGCATGGCTTT-3' and 5'-CACCTTCA-CCGTTCCAGTTT-3'. The standard curve method was used for the quantification of the relative amounts of gene expression products. This method provides unit-less normalized expression values that can be used for direct comparison of the relative amounts of target DNA in different samples. We considered more than 1.5 fold as an increased DNA copy number.
Nine gastric cancer cell lines were cultured at 37℃ in 5% CO2 in Dulbecco's Modified Eagle's Medium with 10% heat-inactivated fetal bovine serum and were harvested 48 hours later. Twenty-four frozen gastric cancer tissues were ground to very fine powder in liquid nitrogen. These samples were suspended in an ice-cold Nonidet P-40 lysis buffer (10 mM HEPES [pH 7.4], 142.5 mM KCl, 0.2% Nonidet P-40, 5 mM EGTA) supplemented with 1 mM dithiothreitol, 12.5 mM β-glycerophosphate, 1 µM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, and 1× protease inhibitor mix (Roche Molecular Biochemicals). Cell lysates (40 µg total proteins) were separated on 8% sodium dodecyl sulfate polyacrylamide gel electrophoresis and blotted onto a Hybond-Polyvinylidene fluoride transfer membrane (Amersham), which had been subsequently proved with anti-UQCRFS1 (Abcam, Cambridge, UK), and then incubated with secondary antibodies conjugated with horseradish peroxidase. The protein bands were detected using enhanced chemiluminescence Western blotting detection reagents (Amersham Pharmacia Biotech, Buckinghamshire, UK). Equal loading of proteins was confirmed by immunoblotting with an anti-β-actin and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody.
One hundred eighty six formalin-fixed paraffin-embedded gastric cancer specimens were examined for UQCRFS1 immunostaining. Histologically, 61 cases were the intestinal-type, 93 were the diffuse-type and 32 were the mixed-type of gastric cancer. The mean size of the tumors was 6.5 cm and 103 cases were located in the middle or lower section of the stomach. Two pathologists screened the histological sections and selected areas with representative tumor cells. Two and one tissue core samples from each cancer and a non-cancerous gastric mucosal area were taken and placed in a new recipient paraffin block using a commercially available microarray instrument (Beecher Instruments, Micro-Array Technologies, Silver Spring, MD, USA), according to established methods.(10)
For immunohistochemical analysis, tissue microarray recipient blocks were constructed containing 186 gastric cancer tissues from formalin-fixed paraffin embedded specimens. To maximize the immunohistochemistry signal, two strategies were used: antigen retrieval in citrate buffer and signal amplification with biotinylated tyramide, as previously described.(11) Two micrometer sections were incubated overnight at 4℃ with UQCRFS1 (1/100; Abcam) antibody. Detection was carried out using biotinylated goat anti-rabbit antibodies (Sigma, St. Louis, MO, USA), followed by incubation with a peroxidase-linked avidin-biotin complex. Diaminobenzidine was used as a chromogen, and the slides were counterstained with Mayer's hematoxylin. The specificity for anti-UQCRFS1 antibodies was confirmed in nine cancer cell lines by Western blot analysis. Staining for the UQCRFS1 antigen was determined as positive when positive staining in cancer cells was stronger than that of corresponding non-cancerous mucosa. Two pathologists reviewed the results independently. As negative controls, the slides were treated with replacement of the primary antibody by non-immune serum.
ArrayCGH was successfully performed in 23 gastric cancer cases. To identify tumor-specific genomic alterations, we normalized the gastric cancer profiles against the matched gastric mucosae and copy number changes, which were found in less than three cases, were excluded. Loci with amplification were defined as a mean test/reference ratio above 1.5 and loci with a test/reference ratio below 0.5 were considered as losses. From twenty-three gastric cancers, we found common copy number gains in 48 different loci and common copy number losses in 19 different loci. Frequently amplified chromosomal regions included 1p, 2q, 3pq, 4q, 5pq, 6pq, 7pq, 8pq, 9pq, 10q, 11pq, 12q, 14q, 15q, 17q, 18q, 19q, 20pq, and 22q, and frequently deleted chromosomal region included 1p, 2q, 3q, 4q, 7p, 9p, 16p, 19pq, and 22q. These results are highly concordant with previous CGH analysis of gastric cancer.(12,13) Among the amplified genes were several oncogenes previously known to be amplified in gastric cancer including ERBB2/HER2, c-myc and CCND1.(14,15)
Of them, gain at 19q12 locus, which the UQCRFS1 gene is located, was found in 5 (21.7%) of 23 gastric cancer tissues and normalized test/reference ratio was above 2.0. Thus, we selected the UQCRFS1 gene as a novel target gene associated with the development or progression of gastric cancer (Fig. 1). Furthermore, amplification of the UQCRFS1 was confirmed by quantitative real-time PCR and increased copy number of UQCRFS1 was detected in 5 (20%) of 25 gastric cancer DNAs (Fig. 2).
A weak to moderate immunopositivity for UQCRFS1 was clearly marked on the cytoplasm and nucleus of corresponding gastric mucosal epithelial cells and some lymphocytes (Fig. 3). However, the surrounding stromal cells such as fibroblasts were negative for UQCRFS1. Overexpression of the UQCRFS1 protein was detected in 105 (56.5%) of the 186 gastric cancers. Interestingly, dysplastic cells in surrounding gastric mucosa also showed strong UQCRFS1 expression (Fig. 3). UQCRFS1 expression was found in 37 (60.6%), 52 (55.9%), and 16 (50.0%) of the following samples: 61 intestinal-type, 93 diffuse-type, and 32 mixed-type gastric cancers, respectively. UQCRFS1 was strongly expressed in 103 (56.6%) of 182 cases with lymph node metastasis. Statistically, there was no significant relationship between UQCRFS1 overexpression and clinicopathologic parameters, including tumor location, tumor size, depth of invasion, histologic type, lymph node metastasis, lymphovascular invasion and 5 year survival rate (Chi-Square test, P>0.05) (Table 1).
The expression of the UQCRFS1 protein was also examined by immunoblotting using total proteins extracted from gastric cancer cell lines and tissues with anti-UQCRFS1 antibody (Fig. 4). Overexpression of the UQCRFS1 was found in 6 (66.7%) of 9 gastric cancer cell lines and 12 (50%) of 24 cancer tissues. Actin and GAPDH levels, as controls for loading, were consistent in these cell lines and tissues.
DNA copy number aberrations are hallmarks of various human cancers. In this study, we investigated DNA copy number changes in 23 gastric cancers by arrayCGH and found frequent chromosomal losses and gains in gastric carcinomas (Table 2, 3). Chromosomal gains at 17q12, which the ERBB2 gene is located, and at 8q22 region containing c-myc gene was frequently observed. Interestingly, gain on chromosome 19q12 locus, which the UQCRFS1 gene is located, was observed in 5 (21.7%) of 23 gastric cancer tissues, including 4 intestinal- and 1 diffuse-type gastric cancers. Since the copy number ratio of test to normal DNA was more than 2 fold at this locus, we considered the UQCRFS1 gene as novel target gene for gastric cancer development or progression. In quantitative real-time PCR, increased copy number of the UQCRFS1 gene was detected in 5 (20%) in 25 gastric cancers. These results suggest that amplification of the UQCRFS1 gene is closely associated with the development of gastric cancer.
Integration of genome-wide arrayCGH and protein expression data has provided new information about the molecular mechanisms of cancer development or progression. For example, up to 40~60% of the amplified genes have showed simultaneous overexpression in primary tumors and cell lines.(16-18) The results of the immunohistochemical experiments showed that overexpression of UQCRFS1 protein was present in 56.5% of the gastric cancers and was demonstrated in the stepwise progression of normal mucosal epithelial cell to dysplastic cells (Fig. 3). Its expression was not associated with the clinical/pathological parameters studied, including tumor location, tumor size and lymph node metastasis, histological type and the 5 year survival (Table 1). In addition, we confirmed overexpression of the UQCRFS1 protein in human gastric cancer cell lines and tissues by Western blot (Fig. 4). According to the report of Kim et al.,(19) two-dimensional electrophoresis proteomics on the mitochondria-enriched fraction revealed high expression of UQCRFS1 in AGS gastric cancer cells. Furthermore, amplification of UQCRFS1 gene was found in breast cancer with high grade of cancer cells, acute myeloid leukemia, and ovarian carcinoma.(20-22) Taken together, our results suggest that overexpression of the UQCRFS1 protein may contribute to the development of gastric cancer.
The UQCRFS1, the Rieske Fe-S protein (RISP), is a key subunit of the cytochrome bc1 complex (complex III) of the mitochondrial respiratory chain.(23,24) This complex generates an electrochemical potential coupled to adenosine triphosphate (ATP) synthesis to transfer electrons from ubiquinol to cytochrome c. Mitochondrial dysfunction can result in excessive production of reactive oxygen species, which stimulate the release of cytochrome c from mitochondrial inter-membrane space to the cytosol.(25) Recently, UQCRFS1/RISP knockdown in breast tumor cell line led to decreased mitochondrial membrane potential as well as a decreased matrigel invasion.(26) Therefore, it is likely that amplification or overexpression of UQCRFS1 may induce mitochondrial oxygen radical formation and rapid proliferation and invasion of cancer cells by rapid synthesis of ATP. However, further functional analysis of the UQCRFS1 gene will certainly broaden our understanding not only of the pathogenesis of gastric cancer but also of the molecular mechanism involved in the mitochondrial respiratory chain in cancer cells.
In conclusion, we investigated DNA copy number changes in 23 gastric adenocarcinomas by arrayCGH and found common gain and loss in 48 and 19 different loci, respectively. Among them, we report here UQCRFS1 gene, which is located at 19q12, as a novel target of gastric cancer development. The UQCRFS1 gene was amplified in 5 (21.7%) of 23 gastric cancers and overexpression of the protein was found in 105 (56.5%) of 186 gastric cancer tissues. In addition, we also confirmed overexpression UQCRFS1 protein in 6 (66.7%) of 9 gastric cancer cell lines and 12 (50%) of 24 gastric cancer tissues by immunoblotting. All of these results suggest that overexpression of the UQCRFS1 gene may contribute to the development or progression of gastric cancers.
Figures and Tables
Fig. 1
Array-comparative genome hybridization result showing amplification of chromosome 19q13 locus, which the UQCRFS1 gene is located, in gastric cancers.
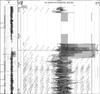
Fig. 2
Copy number change of UQCRFS1 in gastric cancers. In real time polymerase chain reaction, copy number ratio was calculated based on the corresponding normal value of each case. UQCRFS1 copy number was increased more than 1.5 fold in 5 (20%) of 25 gastric cancer DNAs compared to the surrounding gastric mucosal tissue DNAs. Cont = control; T = tumor.
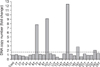
Fig. 3
Immunostaining of the UQCRFS1 protein in gastric cancer. Expression of the UQCRFS1 protein in gastric cancers. Low-power view (×2, magnification) of tissue cores stained for UQCRFS1. Immunostaining for UQCRFS1 was detected in cytoplasm and nucleus of non-cancerous gastric mucosal epithelial cells (×200, magnification) (A, B). Overexpression of UQCRFS1 protein was seen in well differentiated adenocarcinoma (C) and moderate differentiated adenocarcinoma (D) (×200, magnification).
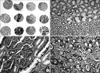
Fig. 4
Immunoblotting analysis of UQCRFS1 protein expression in gastric cancer cell lines and tissues. Total cell lysates prepared from different cell lines and tissues were subjected to immunoblotting analysis with UQCRFS1, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and actin antibodies. Compared with UQCRFS1 expression in corresponding con-cancerous gastric mucosa, expression of the protein was increased in 6 (66.7%) of 9 gastric cancer cell lines and 12 (50.0%) of 24 gastric cancer tissues, respectively.

Acknowledgments
This work was supported by the Happy tech. program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2010-0020764).
References
1. Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, et al. Cancer statistics, 2005. CA Cancer J Clin. 2005. 55:10–30.

2. Jung KW, Park S, Kong HJ, Won YJ, Lee JY, Park EC, et al. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2008. Cancer Res Treat. 2011. 43:1–11.

3. Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, et al. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet. 2001. 29:263–264.

4. Buffart TE, Carvalho B, Hopmans E, Brehm V, Kranenbarg EK, Schaaij-Visser TB, et al. Gastric cancers in young and elderly patients show different genomic profiles. J Pathol. 2007. 211:45–51.

5. Weiss MM, Kuipers EJ, Postma C, Snijders AM, Pinkel D, Meuwissen SG, et al. Genomic alterations in primary gastric adenocarcinomas correlate with clinicopathological characteristics and survival. Cell Oncol. 2004. 26:307–317.

6. Weiss MM, Kuipers EJ, Postma C, Snijders AM, Siccama I, Pinkel D, et al. Genomic profiling of gastric cancer predicts lymph node status and survival. Oncogene. 2003. 22:1872–1879.

7. Sugihara H, Hattori T, Fujita S, Hirose K, Fukuda M. Regional ploidy variations in signet ring cell carcinomas of the stomach. Cancer. 1990. 65:122–129.

8. Hattori T, Sugihara H, Fukuda M, Hamada S, Fujita S. DNA ploidy patterns of minute carcinomas in the stomach. Jpn J Cancer Res. 1986. 77:276–281.
9. Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000. 132:365–386.

10. Kononen J, Bubendorf L, Kallioniemi A, Bärlund M, Schraml P, Leighton S, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998. 4:844–847.

11. Yoon JH, Song JH, Zhang C, Jin M, Kang YH, Nam SW, et al. Inactivation of the Gastrokine 1 gene in gastric adenomas and carcinomas. J Pathol. 2011. 223:618–625.

12. Tsukamoto Y, Uchida T, Karnan S, Noguchi T, Nguyen LT, Tanigawa M, et al. Genome-wide analysis of DNA copy number alterations and gene expression in gastric cancer. J Pathol. 2008. 216:471–482.

13. Rossi E, Klersy C, Manca R, Zuffardi O, Solcia E. Correlation between genomic alterations assessed by array comparative genomic hybridization, prognostically informative histologic subtype, stage, and patient survival in gastric cancer. Hum Pathol. 2011. 42:1937–1945.

14. Bizari L, Borim AA, Leite KR, Gonçalves Fde T, Cury PM, Tajara EH, et al. Alterations of the CCND1 and HER-2/neu (ERBB2) proteins in esophageal and gastric cancers. Cancer Genet Cytogenet. 2006. 165:41–50.

15. Calcagno DQ, Leal MF, Seabra AD, Khayat AS, Chen ES, Demachki S, et al. Interrelationship between chromosome 8 aneuploidy, C-MYC amplification and increased expression in individuals from northern Brazil with gastric adenocarcinoma. World J Gastroenterol. 2006. 12:6207–6211.

16. Wolf M, Mousses S, Hautaniemi S, Karhu R, Huusko P, Allinen M, et al. High-resolution analysis of gene copy number alterations in human prostate cancer using CGH on cDNA microarrays: impact of copy number on gene expression. Neoplasia. 2004. 6:240–247.

17. Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002. 62:6240–6245.
18. Järvinen AK, Autio R, Haapa-Paananen S, Wolf M, Saarela M, Grénman R, et al. Identification of target genes in laryngeal squamous cell carcinoma by high-resolution copy number and gene expression microarray analyses. Oncogene. 2006. 25:6997–7008.

19. Kim HK, Park WS, Kang SH, Warda M, Kim N, Ko JH, et al. Mitochondrial alterations in human gastric carcinoma cell line. Am J Physiol Cell Physiol. 2007. 293:C761–C771.

20. Ohashi Y, Kaneko SJ, Cupples TE, Young SR. Ubiquinol cytochrome c reductase (UQCRFS1) gene amplification in primary breast cancer core biopsy samples. Gynecol Oncol. 2004. 93:54–58.

21. Sait SN, Qadir MU, Conroy JM, Matsui S, Nowak NJ, Baer MR. Double minute chromosomes in acute myeloid leukemia and myelodysplastic syndrome: identification of new amplification regions by fluorescence in situ hybridization and spectral karyotyping. Genes Chromosomes Cancer. 2002. 34:42–47.

22. Kaneko SJ, Gerasimova T, Smith ST, Lloyd KO, Suzumori K, Young SR. CA125 and UQCRFS1 FISH studies of ovarian carcinoma. Gynecol Oncol. 2003. 90:29–36.

23. Rieske JS. Composition, structure, and function of complex III of the respiratory chain. Biochim Biophys Acta. 1976. 456:195–247.

24. Trumpower BL, Edwards CA. Purification of a reconstitutively active iron-sulfur protein (oxidation factor) from succinate. cytochrome c reductase complex of bovine heart mitochondria. J Biol Chem. 1979. 254:8697–8706.





 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download