Abstract
Purpose
Silent mating-type information regulation 2 homologue 1 (SIRT1) is a nicotinamide adenine dinucleotide-dependent deacetylase. SIRT1 plays an important role in the regulation of cell death/survival and stress response in mammals. The aim of this study was to investigate whether the SIRT1 gene is involved in the development or progression of gastric cancers.
Materials and Methods
SIRT1 and p53 genes in 86 gastric cancers were examined for genetic alterations by PCR-single strand conformation polymorphism sequencing, as well as SIRT1 protein expression in 170 gastric cancers by immunohistochemistry.
Results
In the genetic analysis, we found SIRT1 and p53 mutations in two and 12 cases, respectively. Two missense mutations, c.599 C>T (T200I) and c.1258 G>A (E420K), were detected in the SIRT1 gene coding region. The SIRT1 and p53 mutation were found in mutually exclusive gastric cancers. The immunohistochemistry revealed that SIRT1 overexpression was found in 95 (55.9%) of 170 gastric cancers. Altered SIRT1 expression was not statistically associated with clinicopathological parameters, including tumor differentiation, location, lymph node metastasis, or p53 expression. Two cases with an SIRT1 mutation showed increased SIRT1 expression.
Gastric carcinoma is one of the most common carcinomas in the world. In Korea, it accounts for an estimated 20.2% of all malignancies, 24.0% in men and 15.3% in women.(1) At the molecular level, the concept of multistage carcinogenesis is now widely accepted as a consequence of multiple genetic alterations in cancer cells.(2) Oncogene activation and tumor suppressor gene inactivation can cause dysregulated cell growth in gastric cancer.
SIRT1 is one of the seven members of the sirtuin family, and is classified as a Nicotinamide adenine dinucleotide (NAD+)-dependent deacetylase that is critically involved in stress responses, development and cellular metabolism.(3-5) Increased SIRT1 expression in cancer compared to normal cells has been observed in various human malignant tumors, including lymphomas, leukemia and soft tissue sarcomas, prostate cancer, lung and colon carcinomas,(6-8) suggesting that SIRT1 activity is involved in tumorigenesis. The evidence above points to a tumor-promoting role for overactive SIRT1, but other studies clearly showed that it acted as a tumor suppressor by preventing genomic instability.(9,10) Reduction of SIRT1 expression has been detected in cells derived from different tumors, including bladder, prostate carcinomas and ovarian cancers.(10) The controversy over whether SIRT1 serves as a tumor promoter or a tumor suppressor has not been completely resolved and debates will continue.
SIRT1 has deacetylation activity on histones (H1, H3, and H4) and non-histone proteins, including p53, FOXO family proteins, nuclear factor κB and Ku70.(11-14) The tumor suppressor, p53, represents the first non-histone substrate of SIRT1 functionally regulated by acetylation and deacetylation. SIRT1 destabilizes p53 through catalyzing deacetylation of p53 at lysine 382, which weakens its DNA binding ability.(15,16) Accordingly, cells derived from SIRT1 deficient mice and cells treated with siRNAs against SIRT1 show high levels of hyperacetylated p53, and a dominant-negative SIRT1 mutant increases p53-dependent transcriptional activity.(17) However, it is also known that p53 positively regulates SIRT1 transcription and loss of p53 impairs SIRT1 induction.(18) Therefore, it is likely that p53 inactivation can reduce SIRT1 expression level through a negative feedback loop. As such, we elected to examine for possible correlation between SIRT1 and p53 in gastric cancer.
In order to determine whether genetic and expression alterations of the SIRT1 and p53 are involved in the development or progression of gastric cancer, somatic mutations and protein expression of the SIRT1 and p53 genes were analyzed in gastric adenocarcinomas.
One hundred seventy (n=170) formalin-fixed paraffin-embedded gastric cancer specimens were obtained from the College of Medicine, The Catholic University of Korea, between 2003 and 2004. For gastric cancer, 73 cases were the intestinal-type and 97 were the diffuse-type gastric cancer. The mean size of the tumors was 6.5 cm, and 154 cases were found located in the middle or lower section of the stomach. Two pathologists screened the histological sections and selected areas representative of the tumor cells. Two and one tissue core samples from each cancer and normal area were taken and placed in a new recipient paraffin block using a commercially available microarray instrument (Beecher Instruments, Micro-Array Technologies, Silver Spring, MD, USA), according to established methods.(19) The institutional review board of the Catholic University of Korea, College of Medicine (CUMC09U042) approved this study. There was no evidence of familial cancer in patients from which the specimens were obtained.
For the immunohistochemical analysis, 2 µm sections were cut the day before use and stained according to standard protocols. To maximize the signal on immunohistochemistry, two strategies were used in this study: antigen retrieval in citrate buffer and signal amplification with biotinylated tyramide. For the former, heat-induced epitope retrieval was conducted by immersing the slides in Coplin jars filled with 10 mmol/L citrate buffer (pH 6.0) and boiling the buffer for 30 min in a pressure cooker (Nordic Ware, Minneapolis, MN, USA) inside a microwave oven at 700 W. The jars were then cooled for 20 min. For the latter, the Renaissance TSA indirect kit (NEN Life Science, Boston, MA, USA), which included streptavidin-peroxidase and biotinylated tyramide, was used. After rinsing with PBS, the slides were treated with 1% H2O2 in PBS for 15 min at room temperature to abolish endogenous peroxidase activity. After washing with TNT buffer (0.1 mol/L Tris-HCl, pH 7.4, 0.15 mol/L NaCl and 0.05% Tween 20) for 20 min, the slides were treated with TNB buffer (0.1 mol/L Tris-HCl, pH 7.4, 0.15 mol/L NaCl and 0.5% blocking reagent). Sections were incubated overnight at 4℃ with antibodies (1/100 dilution) to SIRT1 protein (Abcam, Cambridge, UK). Detection was carried out using biotinylated rabbit anti-rabbit antibody (Sigma, St. Louis, MO, USA), followed by incubation with peroxidase-linked avidin-biotin complex. Diaminobenzidine was used as a chromogen, and the slides were counterstained with Mayer's hematoxylin. The specificity of anti-SIRT1 antibody was confirmed in 9 cancer cell lines by Western blot analysis (data not shown). Staining for SIRT1 antigen was determined as positive when more than 30% of nucleus was positively stained. Two pathologists reviewed the results independently. As negative controls, the slides were treated with primary antibody replacement by non-immune serum.
Since DNA extracted from formalin fixed paraffin embedded tissue is of low quality for mutational analysis, we extracted DNAs from 86 methacarn-fixed paraffin-embedded tissues, which were not included in our tissue microarray. These samples were also obtained from the College of Medicine, The Catholic University of Korea, between 2003 and 2004. Tumor cells were selectively procured from Hematoxylin & Eosin stained slides using a laser microdissection device (ION LMD, JungWoo International Co, Seoul, Korea). The surrounding normal gastric mucosal cells were also obtained to study the corresponding normal DNA from the same slides in all cases. DNA extraction was performed using a modified single step DNA extraction method, as described previously.(20)
Genomic DNAs from each cancer cell and the corresponding non-cancerous gastric mucosal tissues were amplified with primers covering the entire exons of the SIRT1 and DNA binding domain (exon 4~9) of the p53 gene. Primer sequences are shown in Table 1. Numbering of the sequences of SIRT1 was done with respect to the ATG start codon according to the genomic sequence obtained from Genbank accession no. NM_012238. All cases were screened by SSCP analysis of each exon for the presence of an aberrant band in the tumor DNA compared to normal DNA. Each polymerase chain reaction (PCR) procedure was performed under standard conditions in a 10 µl reaction mixture containing 1 µl of the template DNA, 0.5 µM of each primer, 0.2 µM of each deoxynucleotide triphosphate, 1.5 mM MgCl2, 0.4 unit of Ampli Taq gold polymerase (Perkin-Elmer, Foster City, CA, USA), 0.5 µCi of [32P]dCTP (Amersham, Buckinghamshire, UK), and 1 µl of 10X buffer. The reaction mixture was denatured at 94℃ for 12 min and then incubated for 35 cycles (denaturing at 94℃ for 40 s, annealing at 52~62℃ for 40s and extension at 72℃ for 40s). A final extension step at 72℃ was performed for 5 min. After amplification, the PCR products were denatured for 5 min at 95℃ in a 1 : 1 dilution of sample buffer containing 98% formamide/5 mmol/L NaOH. These products were loaded onto a SSCP gel (FMC Mutation Detection Enhancement system; Intermountain Scientific, Kaysville, UT) containing 10% glycerol. After electrophoresis, the gels were transferred to 3 MM Whatman paper and dried. Autoradiography was then performed using Kodak X-OMAT film (Eastman Kodak, Rochester, NY, USA). DNAs showing mobility shifts were cut out from the dried gels and amplified for 35 cycles using the same primer set. Sequencing of the PCR products was sequenced on the ABI prism 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Moderate to strong immunopositivity for SIRT1 was clearly marked on the nucleus of gastric cancer cells (Fig. 1). However, the corresponding normal gastric mucosa and surrounding stromal cells, including fibroblasts, were focal and weak positive for SIRT1 protein. Aberrant expression of SIRT1 protein was detected in 95 (55.9%) of 170 gastric cancers. SIRT1 expression was found in 47 (64.3%) and 48 (49.5%) of 73 intestinal-type and 97 diffuse-type gastric cancers, respectively. Statistically, there was no significant relationship between SIRT1 protein expression and the clinicopathologic parameters, including differentiation, tumor location, tumor size, lymph node metastasis, lymphatic invasion and vein invasion (Chi-Square test, P>0.05) (Table 2).
In our previous report, moderate to strong nuclear staining for p53 protein was found in 79 (42.5%) of 186 cancers.(21) Interestingly, 29 cases were immunopositive for both SIRT1 and p53 proteins. 50 cases with p53 nuclear staining were negative for SIRT1, and 45 cases demonstrated negative staining for both proteins. Statistically, SIRT1 protein expression was not significantly associated with p53 immunostaining (Chi-Square test, P=0.3366).
The presence of any mutations, possibly associated with aberrant SIRT1 expression, was examined using PCR-based SSCP and sequencing analysis. Expectedly, we found SIRT1 and p53 mutations in 2 and 12 cases, respectively. As shown in Fig. 2 and Table 3, one of the SIRT1 mutations was a missense mutation caused by a single nucleotide substitution, G to A transition at nucleotide 1258. Another SIRT1 mutation was a missense mutation caused by a single nucleotide substitution, C to T transition at nucleotide 599. Two cases showed aberrant bands of the mutant allele with that of the wild type in SSCP analysis, indicating a hemizygous mutation in 1 allele in the presence of the remaining allele. The corresponding normal samples showed no evidence of mutation by repeated SSCP, indicating that the mutations occurred somatically. In immunohistochemistry, two cases with SIRT1 mutation showed aberrant increased expression of SIRT1 in the cancer cells (Data not shown).
The experiments, including tissue microdissection, PCR, SSCP and sequencing analysis, were repeated three times to ensure the reliability of the results.
SIRT1 plays an important role in cell survival under genotoxic and oxidative stress through deacetylation of key cell cycle molecules and apoptosis regulatory proteins.(5,11-14,17) Overexpression of SIRT1 can enhance tumor growth and promote cell survival in response to stress and drug resistance.(22) Moreover, inhibition of SIRT1 function induces growth arrest or apoptosis of human cancer cells.(23,24) Therefore, SIRT1, in addition to serving as a prognostic marker, may also provide a target for a novel therapeutic approach for gastric carcinomas.
Our study has shown that a high proportion of gastric carcinomas overexpressed SIRT1. However, there was no significant relationship between SIRT1 protein expression and the clinicopathologic parameters, including differentiation, location, size and lymph node metastasis, lymphatic invasion and venous invasion. These results did not provide direct evidence that SIRT1 expression is increased with gastric cancer progression. In addition, the presence of any mutations, possibly associated with aberrant SIRT1 expression, was examined using PCR-based SSCP and sequencing analysis. Expectedly, we found two somatic missense mutations of the SIRT1 gene in 86 sporadic gastric cancers. Interestingly, all of the samples with mutations demonstrated increased expression of SIRT1 in cancer cells. These results suggest that SIRT1 overexpression might contribute to the development of a subset of gastric cancers.
The p53 protein is a key regulator of cell cycle progression and apoptosis. Lys382 of p53 is a substrate for the SIRT1 mediated deacetylation, which antagonizes p53-dependent transcriptional activation and apoptosis in response to DNA damage and oxidative stress.(11) During tumor development, up-regulated SIRT1 allows cells to bypass apoptosis and survive DNA damage by deacetylation and p53 inactivation.(15,16) In the present study, we found p53 mutations in 12 (14.0%) of 86 gastric cancers and mutations of SIRT1 and p53 were mutually exclusive. Mutation of p53 is one of the most prevalent genetic alterations in human cancer, including gastric carcinoma. The frequency of p53 mutations in Japanese gastric cancer is at around 40%.(25) Thus, our results may have underestimated the prevalence of the SIRT1 and p53 somatic mutations in gastric cancers, as the sensitivity rate of SSCP analysis for the detection of single-base substitutions is estimated to be only 80%.(26) However, since we repeated the experiments three times, the chance that we missed any mutation is very low. These evidences imply the existence of cultural and environmental factors that can influence p53 mutation between different ethnic groups. Another possibility is that a high frequency of p53 mutation previously reported may be due to formalin fixation of archival specimens.(27) Further studies are very necessary to verify these initial observations.
Next, we have also examined for possible correlation between SIRT1 and p53 expression status. In our previous report, we found p53 nuclear expression in 79 (42.5%) of 186 gastric cancers.(21) In this study, when we compared SIRT1 over-expression with p53 nuclear expression, there was no significant association between p53 and SIRT1 expression (P=0.3366). Therefore, it is possible that SIRT1 in gastric cancer may not be dependent on functional p53. Thus, our results do not support the positive findings of another Korean research group, which reported a significant association between SIRT1 and p53 expression in gastric cancers.(28) Since SIRT1 was known as an exclusively nuclear protein,(29) we considered only nuclear localization as immunopositive expression. Difference in immunopositive criteria is likely to account for the discrepancy. There may be other cellular targets of SIRT1 that regulate dysregulated cancer cell growth, such as nuclear factor κB, Ku70 and Foxo transcription factors as reported previously.(12-14)
SIRT1 has well established anti-apoptotic activity and is presumed to act as an oncogene. However, a recent study showed that SIRT1+/+ mice with Apc mutation inhibit intestinal tumorigenesis.(30) SIRT1-/- mice showed increased tumor incidence when crossed to a p53-/- background,(10) suggesting that SIRT1 has properties of an atypical tumor suppressor. It is likely that SIRT1 plays different functions, depending on different SIRT1 down-stream targets and factors.
Here, we found frequent nuclear overexpression of SIRT1 protein, and 2 and 12 somatic mutations of SIRT1 and p53 genes, respectively. Overexpression of SIRT1 protein was not associated with clinicopathologic parameters and p53 nuclear expression. The cases carrying SIRT1 mutation showed overexpression of SIRT1 protein and mutations of SIRT1 and p53 were mutually exclusive. Although we did not perform functional analysis of SIRT1 mutations in this study, our results suggest that aberrant expression and genetic alterations of SIRT1 may contribute to the development of gastric cancer. Additional studies are needed to clarify the role of the SIRT1 in the pathogenesis of gastric cancers.
Figures and Tables
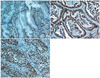 | Fig. 1Aberrant SIRT1 expression in gastric cancer cells. Normal gastric mucosa exhibited focal weak positive staining for SIRT1 (A). Intestinal-(B) and diff use-type (C) gastric cancers displayed nuclear staining for SIRT1 (Original magnification, ×200). |
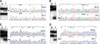 | Fig. 2Representative mutations of the SIRT1 gene (A & B) and p53 gene (C & D) detected in gastric cancer. SSCP demonstrating aberrant bands (arrow) and sequencing data showing missense mutations: (A) c.599 C>T (T200I), (B) c.1258 G>A (E420K) in the SIRT1 gene, (C) c.488A>G (Y163C), (D) c.440T>G (V147G) in the p53 gene (N = non-neoplastic DNA; T = tumor DNA). |
References
1. Shin HR, Jung KW, Won YJ, Park JG. 139 KCCR-affiliated Hospitals. 2002 annual report of the Korea Central Cancer Registry: based on registered data from 139 hospitals. Cancer Res Treat. 2004. 36:103–114.

2. Yuasa Y. Control of gut differentiation and intestinal-type gastric carcinogenesis. Nat Rev Cancer. 2003. 3:592–600.

3. Bordone L, Cohen D, Robinson A, Motta MC, van Veen E, Czopik A, et al. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007. 6:759–767.

4. Murayama A, Ohmori K, Fujimura A, Minami H, Yasuzawa-Tanaka K, Kuroda T, et al. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008. 133:627–639.

5. Hasegawa K, Yoshikawa K. Necdin regulates p53 acetylation via Sirtuin1 to modulate DNA damage response in cortical neurons. J Neurosci. 2008. 28:8772–8784.

6. Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007. 23:413–418.

7. Fraga MF, Agrelo R, Esteller M. Cross-talk between aging and cancer: the epigenetic language. Ann N Y Acad Sci. 2007. 1100:60–74.

9. Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008. 135:907–918.

10. Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008. 14:312–323.

11. Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, et al. hSIR2 (SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001. 107:149–159.

12. Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004. 303:2011–2015.

13. Lee JH, Song MY, Song EK, Kim EK, Moon WS, Han MK, et al. Overexpression of SIRT1 protects pancreatic beta-cells against cytokine toxicity by suppressing the nuclear factor-kappaB signaling pathway. Diabetes. 2009. 58:344–351.

14. Cohen HY, Lavu S, Bitterman KJ, Hekking B, Imahiyerobo TA, Miller C, et al. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol Cell. 2004. 13:627–638.

15. Langley E, Pearson M, Faretta M, Bauer UM, Frye RA, Minucci S, et al. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002. 21:2383–2396.

16. Luo J, Li M, Tang Y, Laszkowska M, Roeder RG, Gu W. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci U S A. 2004. 101:2259–2264.

17. Ford J, Jiang M, Milner J. Cancer-specific functions of SIRT1 enable human epithelial cancer cell growth and survival. Cancer Res. 2005. 65:10457–10463.

18. Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004. 306:2105–2108.

19. Kononen J, Bubendorf L, Kallioniemi A, Bärlund M, Schraml P, Leighton S, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998. 4:844–847.

20. Lee JY, Dong SM, Kim SY, Yoo NJ, Lee SH, Park WS. A simple, precise and economical microdissection technique for analysis of genomic DNA from archival tissue sections. Virchows Arch. 1998. 433:305–309.

21. Kim CJ, Song JH, Cho YG, Cao Z, Kim SY, Nam SW, et al. Activation-induced cytidine deaminase expression in gastric cancer. Tumour Biol. 2007. 28:333–339.

22. Jang KY, Hwang SH, Kwon KS, Kim KR, Choi HN, Lee NR, et al. SIRT1 expression is associated with poor prognosis of diffuse large B-cell lymphoma. Am J Surg Pathol. 2008. 32:1523–1531.

23. Ford J, Jiang M, Milner J. Cancer-specific functions of SIRT1 enable human epithelial cancer cell growth and survival. Cancer Res. 2005. 65:10457–10463.

24. Ota H, Tokunaga E, Chang K, Hikasa M, Iijima K, Eto M, et al. Sirt1 inhibitor, Sirtinol, induces senescence-like growth arrest with attenuated Ras-MAPK signaling in human cancer cells. Oncogene. 2006. 25:176–185.

25. Uchino S, Noguchi M, Ochiai A, Saito T, Kobayashi M, Hirohashi S. p53 mutation in gastric cancer: a genetic model for carcinogenesis is common to gastric and colorectal cancer. Int J Cancer. 1993. 54:759–764.

26. Sheffield VC, Beck JS, Kwitek AE, Sandstrom DW, Stone EM. The sensitivity of single-strand conformation polymorphism analysis for the detection of single base substitutions. Genomics. 1993. 16:325–332.

27. Williams C, Pontén F, Moberg C, Söderkvist P, Uhlén M, Pontén J, et al. A high frequency of sequence alterations is due to formalin fixation of archival specimens. Am J Pathol. 1999. 155:1467–1471.

28. Cha EJ, Noh SJ, Kwon KS, Kim CY, Park BH, Park HS, et al. Expression of DBC1 and SIRT1 is associated with poor prognosis of gastric carcinoma. Clin Cancer Res. 2009. 15:4453–4459.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download