Abstract
Objectives
One of the most important functions for a medical practitioner while treating a patient is to study the patient's complete medical history by going through all records, from test results to doctor's notes. With the increasing use of technology in medicine, these records are mostly digital, alleviating the problem of looking through a stack of papers, which are easily misplaced, but some of these are in an unstructured form. Large parts of clinical reports are in written text form and are tedious to use directly without appropriate pre-processing. In medical research, such health records may be a good, convenient source of medical data; however, lack of structure means that the data is unfit for statistical evaluation. In this paper, we introduce a system to extract, store, retrieve, and analyse information from health records, with a focus on the Indian healthcare scene.
Methods
A Python-based tool, Healthcare Data Extraction and Analysis (HEDEA), has been designed to extract structured information from various medical records using a regular expression-based approach.
Most of the medical records we have today are either in unstructured or semi-structured form. Documents from the domain of laboratory reports, for example, consist of attributes from a closed set of attribute types and their respective measurements, and they are already in the desired structured form. However, data in documents like doctor's notes, discharge statements, etc. is mostly unstructured, and very difficult to analyse.
Healthcare Data Extraction and Analysis (HEDEA) is a system to extract information from clinical reports, including prescriptions, blood and urine test reports, and medical notes. The system stores this information and retrieves it as a single-page anamnesis of the patient, with all past records displayed in a comprehensive tabular form. This can help a physician to thoroughly analyse the patient's medical history without sifting through years of paperwork, some of which go missing.
Another area in which a structured information system can be useful is biomedical research. Epidemiological research is highly dependent on the statistical evaluation of medical data; hence, it requires data available in a structured form [1]. Obtaining such structured medical data through study-specific tests on a carefully selected subgroup of patients is both time and cost-intensive. Another way to obtain the necessary data is to manually examine archives filled with clinical reports and extract the required information, which is also an immensely time-consuming process. It would be desirable to perform analysis of those clinical records automatically or at least semi-automatically.
Other advantages of such a system include easier knowledge transfer between doctors, accountability and compliance of hospital procedures, systematic tracking of patient's health, and preservation of knowledge of current treatments and medical procedures for future reference.
Harkema et al. [2] designed a system to extract key information from clinical and biomedical text. Fette et al. [3] described techniques to process semi-structured clinical data and storage in a data warehouse. Atzmueller et al. [4] showed that a structured information system can help augment the data of an existing medical study or help create completely new complex medical hypotheses by studying patterns in what may seem to be independent factors. Black [5] explained how the data from such a system can be used to check a medical hypothesis. The system can also help determine groups of patients suitable for becoming part of a new study based on selection criteria, as shown by Kamal et al. [6]. Concerns related to doctor–patient confidentiality breaches were addressed to some extent by the systems designed by Aberdeen et al. [7] and Yang and Garibaldi [8]. Information extraction using various machine learning models has also been explored by Sondhi et al. [9], Uzuner et al. [10], Bae and Kim [11], Park et al. [12] and Glavas [13], and further analysed by Kraus et al. [14]. Chang et al. [15] proposed an approach for auto-assessment of health quality using classification and identification techniques.
While similar systems do exist for specific applications in various parts of the world, there is a need for system tailored for use in Indian healthcare. Athavale and Zodpey [16] discussed the importance of informatics in the context of Indian healthcare and highlighted the lack of effective Electronic Health Records (EHR) management and analytics, while suggesting potential benefits of EHR in India. Another shortcoming of previously proposed systems is that they mostly cater to one specific requirement. Since most of the data is the same, multiple systems for different use cases and users (patients, physicians, and researchers), cause unnecessary duplication, making maintenance of databases difficult and cumbersome. There is a need to design a comprehensive system that caters to all kinds of users through different access modes, while maintaining data consistency and privacy. We tried to solve this problem through HEDEA, as proposed in this paper.
The complete system involves three primary units—an information extraction unit, an information storage unit, and an analysis unit. The information extraction unit extracts information from standard semi-structured text files using regular expressions, and passes it to the information storage unit, which stores the information under relevant attributes in an encrypted centralized database using the patient ID information as the primary key. This database is then queried by the analysis unit to generate results.
For identification of a patient across healthcare facilities, we use the national identification system in India called Aadhaar, a 12-digit number issued to citizens of the country. Using this identification number coupled with biometric authorisation, the patient can allow access to his/her medical history to the physician as and when needed, or it is possible for the patient to lock it to prevent any unauthorised access of information. If the information is locked by the patient, it cannot even be used for anonymous analytics.
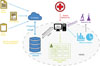
The functionality architecture of HEDEA is shown in Figure 1. The complete data is stored in a centralized database, accessible over the internet via appropriate user access keys.
Users upload medical reports to the system, where relevant information is extracted by the information extraction unit using regular expressions, tagged and stored by the information storage unit in the patient accounts, and based on access rights assigned by the patient, a comprehensive analytics database (CAD) for analysis and research without identifying information. The system uses regular expressions because most of the information to be extracted follows a precise format and resides in the vicinity of identifying information tags. Regular expressions make it easier to identify such information, and they vastly reduce the prospect of noise in the extracted information.
For any query related to a patient's medical history, the patient's Aadhaar number is fed to the system's analysis & output (A&O) unit, authenticated by the patient using a password, and the patient's complete (or partial, controlled by ‘from’ and ‘to’ dates) medical record is presented. For any query in the category of research & analysis, the query is fed into the A&O unit, and the results for the category are presented accordingly in raw format or in the form of charts or graphs, as needed. The data in this case is sourced from the CAD, which does not contain any identifying information.
The information extraction unit was written in Python 3, and it uses regular expressions to extract information from text with a specified format. Data corresponding to attributes such as date of examination, weight, height, symptoms, and prescribed medicine are extracted from the file and stored along with the patient's ID number in a file for each visit. Similarly, blood and urine test results with the date and patient ID are also extracted and stored in the database.
The information set used to design the information extraction unit was created by collecting a set of 90 healthcare reports from different patients with their consent. It contains a mix of doctors' notes, blood and urine test reports, and prescription files, from a number of healthcare institutes.
The input text files are scanned for matching patterns, and the relevant information is extracted. This is done with the help of regular expressions. For semi-structured data, the regular expressions directly extract the required data because the relevant data is expected to be labelled to a reference keyword, albeit in different formats. The data value should follow a specified format or type, or it should be in the vicinity of a reference keyword, as defined in the list of regular expressions.
For unstructured data, generally found in discharge notes and the like, another approach using regular expressions is used. A data value in the unstructured text should follow a specified format or type, or it should be a part of a list of keywords. As an example, for the reference keyword ‘blood pressure’, keywords such as ‘high’, ‘low’, or ‘normal’ are allowed, or numerical strings such as ‘110/90’ or ‘120/80’. The data value should lie in the vicinity of the reference keyword. Each probable data value is assigned a score based on the distance from the keyword, which is the difference between the total number of words in the sentence with the keyword and the number of words occurring between the two. The score is reduced by a large factor if the keyword and data value occur in different sentences. The score is augmented slightly for numerical data values. For example, as shown in Figure 2, ‘120/80’ will have a higher score than the word ‘normal’. Thus, the attribute ‘blood pressure’ is assigned a value ‘120/80’ in the database for the particular tuple.
Some extracted information, such as date and ID information, are processed before storage to maintain consistency in data formats, thus making operations like searching and sorting of data easier.
The output is obtained in an HTML page generated by the HEDEA A&O unit, which was developed using common gateway interface (CGI) scripting. The output, formatted in tables, has attributes as columns and corresponding records with dates as rows. Thus, any fluctuation in a patient's vital signs and test results can be easily detected, and relevant medical action can be taken. The page shows results in three categories, namely, prescriptions, blood reports, and urine reports. An example output has been shown in Figure 3. This is now a completely structured transformation of the unstructured data available in the reports.
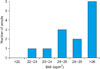
Patient history can also be analysed using charts that can be generated on any numerical attribute. This has been demonstrated using the blood sugar levels of a patient over four weeks in Figure 4. HEDEA enables data aggregation and visualization even if the tests were performed at different pathological laboratories.
HEDEA can be used to generate analysis reports and charts using information in the central database without identifying information of the patient. This information is only provided if the patient allows the information to be used for medical research purposes by allowing the said information to be stored into the CAD. An example chart is shown in Figure 5. This chart uses the calculated body mass index (BMI) of all patients with relevant details in the CAD using the latest weight and height information, and presents the count of people falling in each segment.
In this paper, we introduced an information extraction and presentation system that was designed to recognize and classify basic attributes present in medical records. We proposed a natural language processing model involving keyword-based and rule-based approaches to cope with the inherent complexity and structure of these records. A rich set of features are extracted using regular expression template patterns. At the retrieval step, only the necessary information is displayed for the relevant user, with all personal information removed from data for medical research, and only patient-authorised information available to the medical practitioner, with authentication based on Aadhaar ID.
For data extraction from unstructured text, an additional layer of model-based search using a convolutional neural network can be used to verify obtained results. This has been demonstrated by Li and Huang [17] for this exact use case.
A mobile application can be designed for easier access to patients in comparison to the current web-based solution, which will help encourage adoption of the system.
Figures and Tables
References
1. Dinu V, Nadkarni P. Guidelines for the effective use of entity-attribute-value modeling for biomedical databases. Int J Med Inform. 2007; 76(11-12):769–779.

2. Harkema H, Roberts I, Gaizauskas R, Hepple M. Information extraction from clinical records. In : Proceedings of the 4th UK e-Science All Hands Meeting; 2005 Sep 19–22; Nottingham, UK.
3. Fette G, Ertl M, Worner A, Kluegl P, Stork S, Puppe F. Information extraction from unstructured electronic health records and integration into a data warehouse. In : Proceedings of the 57th Annual Meeting of the German Society for Medical Informatics, Biometry and Epidemiology (GMDS); 2012 Sep 16–20; Braunschweig, Germany. p. 1237–1251.
4. Atzmueller M, Beer S, Puppe F. A data warehouse-based approach for quality management, evaluation and analysis of intelligent systems using subgroup mining. In : Proceedings of the 22nd International Florida Artificial Intelligence Research Society Conference (FLAIRS); 2009 May 19–21; Sanibel Island, FL. p. 372–377.
5. Black N. Why we need observational studies to evaluate the effectiveness of health care. BMJ. 1996; 312(7040):1215–1218.

6. Kamal J, Pasuparthi K, Rogers P, Buskirk J, Mekhjian H. Using an information warehouse to screen patients for clinical trials: a prototype. AMIA Annu Symp Proc. 2005; 2005:1004.
7. Aberdeen J, Bayer S, Yeniterzi R, Wellner B, Clark C, Hanauer D, et al. The MITRE Identification Scrubber Toolkit: design, training, and assessment. Int J Med Inform. 2010; 79(12):849–859.

8. Yang H, Garibaldi JM. Automatic detection of protected health information from clinic narratives. J Biomed Inform. 2015; 58 Suppl. S30–S38.

9. Sondhi P, Gupta M, Zhai C, Hockenmaier J. Shallow information extraction from medical forum data. In : Proceedings of the 23rd International Conference on Computational Linguistics: Posters; 2010 Aug 23–27; Beijing, China.
10. Uzuner O, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc. 2010; 17(5):514–518.

11. Bae I, Kim JS. A refinement system for medical information extraction from text-based bilingual electronic medical records. J Korean Soc Med Inform. 2008; 14(3):267–274.

12. Park YT, Lee YT, Jo EC. Constructing a real-time prescription drug monitoring system. Healthc Inform Res. 2016; 22(3):178–185.

13. Glavas G. TAKELAB: medical information extraction and linking with MINERAL. In : Proceedings of the 9th International Workshop on Semantic Evaluation (SemEval); 2015 Jun 4–5; Denver, CO. p. 389–393.
14. Kraus S, Blake C, West SL. Information extraction from medical notes. In : Proceedings of the 12th World Congress on Health (Medical) Informatics: Building Sustainable Health Systems; 2007 Aug 20–24; Brisbane, Australia. p. 1913–1915.
15. Chang P, Huang FP, Lai ML. The feasibility of using classification and identification techniques to auto-assess the quality of health information on the web. J Korean Soc Med Inform. 2009; 15(3):247–254.

16. Athavale AV, Zodpey SP. Public health informatics in India: the potential and the challenges. Indian J Public Health. 2010; 54(3):131–136.

17. Li P, Huang H. Clinical information extraction via convolutional neural network [Internet]. Ithaca (NY): arXiv.org;2016. cited at 2018 Apr 1. Available from: https://arxiv.org/abs/1603.09381.




 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download