Abstract
Objectives
A distributed research network (DRN) has the advantages of improved statistical power, and it can reveal more significant relationships by increasing sample size. However, differences in data structure constitute a major barrier to integrating data among DRN partners. We describe our experience converting Electronic Health Records (EHR) to the Observational Health Data Sciences and Informatics (OHDSI) Common Data Model (CDM).
Methods
We transformed the EHR of a hospital into Observational Medical Outcomes Partnership (OMOP) CDM ver. 4.0 used in OHDSI. All EHR codes were mapped and converted into the standard vocabulary of the CDM. All data required by the CDM were extracted, transformed, and loaded (ETL) into the CDM structure. To validate and improve the quality of the transformed dataset, the open-source data characterization program ACHILLES was run on the converted data.
Results
Patient, drug, condition, procedure, and visit data from 2.07 million patients who visited the subject hospital from July 1994 to November 2014 were transformed into the CDM. The transformed dataset was named the AUSOM. ACHILLES revealed 36 errors and 13 warnings in the AUSOM. We reviewed and corrected 28 errors. The summarized results of the AUSOM processed with ACHILLES are available at http://ami.ajou.ac.kr:8080/.
A distributed research network (DRN) enables observational studies to be conducted using multiple data sources, while confidential personal health data remain with the original data holders [1]. A DRN can provide network-wide results by running the same analysis program for participating organizations using the same data structure, called a Common Data Model (CDM), and then combining the summarized results through the network [1]. Research collaborations, including the Observational Health Data Sciences and Informatics (OHDSI), the Observational Medical Outcomes Partnership (OMOP), and Mini-Sentinel project, have proposed DRNs [234].
By providing all of their work products as open-source, OHDSI lowered the technical barriers required for participation in a DRN [2]. However, differences in data structures and coding system are still major barriers to being a data partner in a DRN. Most hospitals in Korea use Electronic Health Record (EHR) systems developed in-house, rather than off-the-shelf EHR systems. Furthermore, many of Korean codes for diagnosis, drugs, and procedures are not compatible with international coding systems.
The adoption and use of EHRs has been increasing worldwide, but most EHRs are not interchangeable [56]. Recently, we converted our 20 years of EHR data to CDM ver. 4, and can now run open-source software that fits the CDM, enabling us to join several international studies instantly. We believe that sharing our experience of converting EHR data to the CDM can serve as a blueprint for other researchers who want to transform their data to the CDM, and could facilitate data holders' participation in a DRN.
This study describes our conversion of EHR data to CDM ver. 4. For this conversion, we mapped codes from local coding systems into the standard vocabulary of OHDSI, performed data conversion called 'extraction, transformation, and loading (ETL)', and checked and improved the data quality. To validate our conversion, we ran the data characterization program Automated Characterization of Health Information at Large-scale Longitudinal Evidence Systems (ACHILLES) using the converted data.
The hospital was a Korean tertiary teaching hospital with 1,096 patient beds and 23 operating rooms that adopted a computerized provider order entry (CPOE) system in 1994 and a comprehensive EHR system in March 2010.
To standardize the format and content of observational data, CDM ver. 4.0 of OMOP was released in April 2012 [4]. The CDM contains 18 data tables: Person, Drug Exposure, Drug Era, Condition Occurrence, Condition Era, Observation Period, Observation, Procedure Occurrence, Visit Occurrence, Death, Drug Cost, Procedure Cost, Location, Provider, Organization, Care Site, Payer Plan Period, and Cohort. OMOP defined a standardized vocabulary and requires the use of that vocabulary in the CDM. Now, OHDSI supports and updates the OMOP CDM.
Local codes for diagnoses, drugs, procedures, and laboratory tests were mapped into the OMOP standard vocabulary and reviewed by two physicians and two nurses. The coding system used for diagnosis in the subject hospital is the Korean Standard Classification of Diseases ver. 5 (KCD-5), a Korean derivative of the International Statistical Classification of Diseases and Related Health Problems, 10th revision (ICD-10), while the standard vocabulary of OMOP for diagnosis is based on the Systematized Nomenclature of Medicine Clinical Terms (SNOMED-CT). Because there was no mapping table from ICD-10 to SNOMED-CT in the version of the OMOP vocabulary available at the time of this analysis, we created our own mapping. Roughly 3,000 KCD-5 terms matched exact terms in the standard OMOP vocabulary, while the others had to be mapped manually. If there was no exact mapping term in the standard vocabulary, a parent term with broad meaning was mapped instead. As a result, 98.4% of the 20,721 KCD-5 codes were mapped to the CDM standardized vocabularies. Our local drug codes were mapped to the OMOP standardized vocabularies, which use RxNorm and the Anatomical Therapeutic Chemical (ATC) classification system. We could map 75.6% of the 5,233 local drug codes. However, unmapped drug codes were rarely used in our database, and their proportion of total prescription counts was only 0.4%. Of the 8,488 local procedure codes (anesthesia, laboratory tests, pathology, radiology, and surgery), 89.3% were mapped to codes in the OMOP standardized vocabularies, based on the Healthcare Common Procedure Coding System (HCPCS), the ICD 9th revision procedure coding system (ICD-9-PCS), and the Current Procedural Terminology, 4th edition (CPT-4) vocabularies.
The ETL process involves pulling data out of one database system and pushing them into another different database system. Of the 18 tables defined in OMOP CDM ver. 4, we performed the ETL process on all but four tables: Drug Cost, Procedure Cost, Payer Plan Period, and Provider. Because we planned to open our converted data to researchers, some data in the excluded tables were considered too sensitive to be opened. The Payer Plan Period table could not be included because Korean has a single mandatory governmental payer. Detailed documentation of the ETL is available in Supplementary Materials.
The standardized dataset constructed using the above ETL process was named the AUSOM (Ajou University School of Medicine), pronounced 'awesome', database. The AUSOM database contains 2,073,120 individuals, 18,717,764 conditions (diagnoses), 99,331,794 drug exposures, and 15,002,879 procedures. Table 1 lists the baseline characterization of the population in the AUSOM database.
ACHILLES is open-source analytics software produced by OHDSI that runs on OMOP CDM ver. 4 and 5 for data characterization, quality assessment, and the visualization of observational health data [2]. ACHILLES calculates summary statistics securely within each local environment, and then a web interface constructs interactive graphic reports using the summary statistics. ACHILLES includes a unique function for checking data quality, named Achilles Heel. Achilles Heel issued 36 errors and 13 warnings from our initial AUSOM data. We fixed 28 errors, but eight errors related to incomplete code mapping still remained. Detailed description and the correction processes for errors and warnings are given in Supplementary Materials. The ACHILLES web on the AUSOM dataset is available at http://ami.ajou.ac.kr:8080/ (Figure 1).
We successfully converted our EHR to the CDM used within the OHDSI community and provided summary statistics for the data in an interactive webpage.
Controversies among studies of the same topic often arise due to differences in the participants, study designs, or interpretations [789]. Even very large individual databases are insufficient to meet the diverse needs of researchers [10]. In a DRN environment, the same study protocol can be run on many participating data sources, and the results from each data source can be combined and compared, while patient privacy and confidentiality are maintained. A DRN can mitigate or even overcome the lack of statistical power due to rare events or selection bias originating from small local datasets.
OHDSI provides open-source software for not only implementing a CDM but also conducting analyses on a CDM [2]. As shown in the case description, ACHILLES summarizes a data source graphically. The OHDSI community is also actively developing open-source analytics for population-level estimation and patient-level prediction. If a patient-level database conforms to the OMOP CDM, any researcher can freely download and use these programs for his/her research.
In a DRN, one study protocol and the associated analytic code can be shared among data partners [111213]. Because the data structure is the same in each database, an analysis code written in one institute can be reused directly in another institute without any revisions. This allows unlimited replication studies of various data sources simultaneously, allowing more reliable and precise results [5]. However, it is still necessary to understand the characteristics of each data source and the detailed ETL process to conduct a study using a DRN [14]. Usually EHR data has a shorter observation period than that of claim data; however, EHR data has more detailed information, such as vital signs, laboratory test results, and exact times of drug administration.
Two main limitations still exist in our data conversion. First, the code mapping process was imperfect. Two physicians and two nurses reviewed the code mapping, but because of different concepts and granularity between the coding systems, information loss was inevitable. Therefore, we need to revise our code mapping and improve it continuously, and update the AUSOM database accordingly. Second, we did not include cost information. Because we planned to open our data to the public via the ACHILLES webpage, we were reluctant to include such sensitive data. This limits the ability to conduct cost-effectiveness studies at present.
The summary statistics in the AUSOM database are open to the public via http://ami.ajou.ac.kr:8080. Data characterizing the population structure, prevalence of conditions, patterns of drug use, laboratory results, and other parameters are available via a user-friendly interactive interface. This graphic interface will help researchers gain insights into realworld practice.
We successfully converted local EHR data to OMOP CDM ver. 4 and opened its summary statistics to the public. The AUSOM database will be revised and updated continuously. We are ready to share our experience and data with anyone who wishes to adopt the OHDSI DRN.
Figures and Tables
 | Figure 1AUSOM data visualized using ACHILLES (Automated Characterization of Health Information at Large-scale Longitudinal Evidence Systems). (A) Basic information about the population in the database is shown in the dashboard tab. (B, C) The prevalence and number of records per person are shown using the size and color of the boxes in the tree maps at the tops of the following tabs: Conditions, Condition Eras, Observations, Drug Eras, Drug Exposures, Procedures, and Visits. Trends and related information for the selected box in the tree map at each tab are visualized below the tree map. The data for (B) essential hypertension and (C) the serum and plasma cholesterol levels are shown (doughnut chart at the bottom right; blue, above the reference range; orange, below the reference range; green, within reference range). The website is available at http://ami.ajou.ac.kr:8080/. |
Table 1
Demographic and clinical characteristics of the population in the AUSOM database
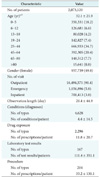
Values are presented as number (%) or the mean ± standard deviation.
aAge at the first observation. bThe difference between the number of persons and the sum of the number of individuals in all age categories is due to missing data in the Observation Period table for some individuals in the source database.
Acknowledgments
This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (No. HI14C3201).
References
1. Platt R, Wilson M, Chan KA, Benner JS, Marchibroda J, McClellan M. The new Sentinel Network: improving the evidence of medical-product safety. N Engl J Med. 2009; 361(7):645–647.

2. Observational Health Data Sciences and Informatics. Analytic tools [Internet]. [place unknown]: Observational Health Data Sciences and Informatics;c2015. cited at 2015 Dec 1. Available from: http://www.ohdsi.org/analytic-tools/.
3. Mini-Sentinel project [Internet]. [place unknown]: Mini-Sentinel Coordinating Center;c2014. cited at 2015 Dec 1. Available from: http://www.mini-sentinel.org/.
4. Observational Medical Outcomes Partnership. OMOP Common Data Model [Internet]. [place unknown]: Observational Medical Outcomes Partnership;2013. cited at 2015 Dec 1. Available from: http://omop.org/CDM.
5. Boyce RD, Ryan PB, Noren GN, Schuemie MJ, Reich C, Duke J, et al. Bridging islands of information to establish an integrated knowledge base of drugs and health outcomes of interest. Drug Saf. 2014; 37(8):557–567.

6. Yoon D, Chang BC, Kang SW, Bae H, Park RW. Adoption of electronic health records in Korean tertiary teaching and general hospitals. Int J Med Inform. 2012; 81:196–203.

7. Madigan D, Ryan PB, Schuemie M, Stang PE, Overhage JM, Hartzema AG, et al. Evaluating the impact of database heterogeneity on observational study results. Am J Epidemiol. 2013; 178(4):645–651.

8. Dodd S. Debating the evidence: oral contraceptives containing drospirenone and risk of blood clots. Curr Drug Saf. 2011; 6(3):132–133.

9. Lockhart PB, Bolger AF, Papapanou PN, Osinbowale O, Trevisan M, Levison ME, et al. Periodontal disease and atherosclerotic vascular disease: does the evidence support an independent association? A scientific statement from the American Heart Association. Circulation. 2012; 125(20):2520–2544.

10. Brown JS, Holmes JH, Shah K, Hall K, Lazarus R, Platt R. Distributed health data networks: a practical and preferred approach to multi-institutional evaluations of comparative effectiveness, safety, and quality of care. Med Care. 2010; 48:6 Suppl. S45–S51.
11. Curtis LH, Weiner MG, Boudreau DM, Cooper WO, Daniel GW, Nair VP, et al. Design considerations, architecture, and use of the Mini-Sentinel distributed data system. Pharmacoepidemiol Drug Saf. 2012; 21:Suppl 1. 23–31.

12. Matcho A, Ryan P, Fife D, Reich C. Fidelity assessment of a clinical practice research datalink conversion to the OMOP common data model. Drug Saf. 2014; 37(11):945–959.

Supplementary Materials
Supplementary materials can be found via http://dx.doi.org/10.4258/hir.2016.22.1.54.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download