Abstract
Objectives
Methyl-CpG binding protein 2 (MeCP2) is a ubiquitous epigenetic factor that represses gene expression by modifying chromatin. Mutations in the MeCP2 gene cause Rett syndrome, a progressive neurodevelopmental disorder. Recent studies also have shown that MeCP2 plays a role in carcinogenesis. Specifically, functional ablation of MeCP2 suppresses cell growth and leads to the proliferation of cancer cells. However, MeCP2's function in adult tissues remains poorly understood. We utilized a weight matrix-based comparison software to identify transcription factor binding site (TFBS) of MeCP2-regulated genes, which were recognized by cDNA microarray analysis.
Methods
MeCP2 expression was silenced using annealed siRNA in HEK293 cells, and then a cDNA microarray analysis was performed. Functional analysis was carried out, and transcriptional levels in target genes regulated by MeCP2 were investigated. TFBS analysis was done within genes selected by the cDNA microarray analysis, using a weight matrix-based program and the TRANSFAC 6.0 database.
Results
Among the differentially expressed genes with a change in expression greater than two-fold, 189 genes were up-regulated and 91 genes were down-regulated. Genes related to apoptosis and cell proliferation (JUN, FOSL2, CYR61, SKIL, ATF3, BMABI, BMPR2, RERE, and FALZ) were highly up-regulated. Genes with anti-apoptotic and anti-proliferative functions (HNRPA0, HIS1, and FOXC1) were down-regulated. Using TFBS analysis within putative promoters of novel candidate target genes of MeCP2, disease-related transcription factors were identified.
Mutations in the MECP2 gene cause Rett syndrome (RTT, OMIM #312750). RTT is an X-linked dominant neurodevelopmental disorder, which is the second major cause of mental retardation in females after Down syndrome [1]. RTT is characterized by the progressive loss of intellectual function, impaired motor skills, deceleration of head growth, and stereotypical hand movements that begin to occur at 6–18 month of age. RTT has an incidence of 1:10,000–1:15,000 female births [2]. Complete absence of functional MeCP2 in males typically results in fatal infantile encephalopathy [3].
MeCP2 is an epigenetic regulator of gene expression [4] that is ubiquitously expressed in almost every tissue, although the level varies by tissue type. MeCP2 expression is elevated in various carcinomas, including mammarygland carcinoma, lymphoma, and colorectal carcinomas [567]. MeCP2 has been reported to function in the chromatin architecture, RNA splicing regulation, and transcriptional activation, as well as DNA methylation-independent gene silencing [8]. However, its function in RTT and neoplastic diseases remains largely unknown. Therefore, to elucidate the exact mechanism of MeCP2 in human diseases, including RTT and cancer, it is important to identify the genes regulated by MeCP2. Transcriptional regulation is one of the most important regulatory mechanism in living cells, and the classification of transcription factor binding domains would enable the prediction of regulatory roles of certain DNA elements [9]. Computational methods of predicting transcription factor binding sites (TFBS) are crucial for understanding the mechanism of gene regulation [10]. We utilized Match, a weight matrix-based comparison software based on position-specific scores to identify TFBS of MeCP2-regulated genes, which were recognized by cDNA microarray analysis [1112].
HEK293 cells were maintained in Dulbecco/Vogt modified Eagle's minimal essential medium (DMEM; Gibco-BRL, Grand Island, NY, USA) containing 10% fetal bovine serum, streptomycin (100 µg/mL), and penicillin (100 units).
hMeCP2 expression was inhibited using annealed siRNA with siPORT Amine (Ambion, Austin, TX, USA) as the transfection agent. The sequences of hMeCP2-specific siRNA were 5'-GGCAAAGCAGAGACAUCAGtt-3' and 5'-CUGAUGUCUCUGCUUUGCCtg-3'.
The total RNA from the HEK293 cells was isolated with TRI Reagent (Gibco-BRL) and purified using RNeasy kits (Qiagen, Valencia, CA, USA). The concentration and integrity of the total RNA were assessed using Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA, USA).
The synthesized cDNA were followed by coupling with Cy3 dye for reference or Cy5 dye for test sample (Amersham). The probes were placed onto the cDNA chip (GenePlorer TwinChip Human-8K; Digital-Genomics, Seoul, Korea) and were hybridized at 58℃.
The chips were scanned with a GenePix 4000A scanner (Axon, Foster City, CA, USA) and were analyzed with GenePix 3.0 software to obtain gene expression ratios. Significance analysis of microarray (SAM) was used to identify the genes with statistically significant changes in expression [13]. A false discovery rate of less than 5% was considered as statistically significant.
To predict the transcriptional regulatory network of the target genes regulated by MeCP2, TFBS analysis was performed. Transcription factors interact with specific elements within the promoter regions of target genes and thus regulate gene expression. TFBS analysis was done within a putative promoter for each up-regulated gene found by the cDNA microarray analyses, with a four-fold increase as the cutoff. Match 1.0, a weight matrix-based program, and the TRANSFAC 6.0 database (BIOBASE International, Wolfenbuettel, Germany) were used for analysis (Figure 1) [914]. Promoters were selected to 1,000 bp downstream of the transcriptional start sites. Through 'vertebrates' matrices with 'minimize the false positive rate' option, the results for several tissue-specific profiles, such as liver, muscle, immunecells, and cell cycle, were merged into a single table. The TFs found for the promoter sequence of each gene were listed, and Pathway Studio (Elsevier Inc., Amsterdam, The Netherlands) was used to study interactions between TFs common to the identified target genes of MeCP2.
To determine the genes repressed by MeCP2, an RNA silencing experiment was carried out using MeCP2-specific siRNA. First, cells were transfected with siRNA-GAPDH as a control to exclude nonspecific effects and cytotoxicity caused by siRNA. GAPDH expression levels were repressed by siRNA, while MeCP2 expression remained stable in HEK293 cells. Cells transfected with amine alone in the absence of siRNA were used as negative controls for MeCP2 expression. The level of MeCP2 mRNA was reduced by 60% in the HEK293 cells treated with siRNA of MeCP2 (siMeCP2), while the HEK293 and amine-treated HEK293 cells showed similar levels of MeCP2 mRNA expression (Figure 2). These results indicate that MeCP2 expression at the RNA and protein levels was efficiently inhibited by siRNA-MeCP2 in the HEK293 cells.
To identify novel target genes of hMeCP2, a cDNA microarray was performed. Of the 8,170 genes tested, 5,215 genes were statistically significant changes and were used for further analyses. Off-target effects caused by siRNA targeting similar sequences in unrelated genes or by global up-regulation of interferon-stimulated genes (ISGs) were excluded from the differential expression profiles [1516], although two of them (DNAJC3 and C2) changed more than two-fold (Table 1). Based on previous reports and gene ontology [15], the differentially expressed genes were grouped according to functions (Table 2).
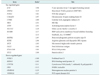
Among the 5,215 genes, 280 genes were differentially expressed in siMeCP2-treated HEK293 cells, with a two-fold difference in expression relative to that in the control HEK293 cells. A total of 189 genes were up-regulated, and 91 genes were down-regulated. The ratio given in Table 3 represents the log2 of Cy5 (siRNA-MeCP2)/Cy3 (control). Table 4 lists the 22 differentially expressed genes with a greater than four-fold change in expression. Four genes (JUN, FOSL2, ATF3, and RERE) were related to apoptosis, and eight genes (JUN, FOSL2, CYR61, SKIL, ATF3, BMABI, BMPR2, and FALZ) were related to cell growth, proliferation, and tumorigenesis. HPA6 responded to cellular stress, and three genes (SCL3A2, MTP, and STC2) showed other functions, including metabolic regulation. Three genes (C18orf25, U45880, and KIAA1340) had no identified function. Furthermore, genes with anti-apoptotic and proliferative functions (HNRPA0, HIS1, and FOXC1) were down-regulated. The target genes of MeCP2 identified by the cDNA microarray were confirmed using quantitative reverse transcriptase-polymerase chain reaction. These results indicate that hMeCP2 modulates the expression of genes related to apoptosis and proliferation.
The up-regulated genes were activated by the siRNA-mediated silencing of the transcriptional repression of MeCP2. Of the 16 genes up-regulated by more than four-fold, five genes (HPA6, RERE, C18orf25, KIAA1340, and U45880) were excluded from the analysis. HPA6 was excluded because its increased expression appeared to be a false-positive effect attributable to a stress response to siRNA. RERE was excluded because it had repeated sequences that made prediction of its promoter difficult. The remaining three genes (C18orf25, KIAA1340, and U45880) were excluded because their functions were unknown. The remaining 11 genes were selected for TFBS analysis. The TFs that matched the MeCP2-regulated genes in the TRANSFAC database are listed in Table 5. The promoters of the selected genes had 12 TFBSs in common; six additional TFBSs were also found to be common to the promoter regions of eight to ten of the genes. The common TFBSs of the newly identified target genes also appeared to be common to the previously validated target genes of MeCP2 (Figure 3).
Previous functional analyses of MeCP2 with regard to RTT have focused exclusively on neurons. Although RTT is primarily a neurodevelopmental disorder, it is also a multisystemic disease that includes abnormalities in the cardiac, respiratory, gastrointestinal, and musculoskeletal systems [17]. Little is known about the target and regulatory genes of MeCP2. Knowledge about the targets of MeCP2 and their functions would be valuable in developing a therapy for RTT patients.
The present study identified genes regulated by MeCP2 in vitro as well as their functions in silico. siRNA-induced RNA interference effectively reduced MeCP2 expression by 66%. Transfection with siRNA-MeCP2 had no cytotoxic effects on HEK293 and was highly specific for the target gene.
Several of the genes up-regulated by RNA silencing of MeCP2 were apoptosis-related genes, including JUN, FOSL2, ATF3, and RERE. Apoptosis plays an important role in several neurodegenerative disorders, such as Huntington disease, Alzheimer disease, Parkinson disease, and amyotrophic lateral sclerosis [18]. JUN and ATF3 are commonly used as markers of neuronal injury [19]. JUN and FOS also induce apoptotic cell death in cancer [20]. The MeCP2 protein is more highly expressed in most cancer tissues than in corresponding noncancerous tissues [21]. The up-regulation of apoptosis-related genes by MeCP2 RNA silencing suggests that the apoptosis-related genes might be suppressed in tumors with high MeCP2 expression. Similarly, decreased expression of apoptosis-related genes may promote tumor overgrowth. It has been suggested that the carcinogenic mechanism of the breast cancer 1 gene may be hypermethylation of apoptosis-related genes [22].
Of the up-regulated genes, RERE and FALZ are reported to be highly expressed in neurodegenerative diseases [2324]. Overexpression of RERE induces apoptosis by recruiting a fragment of the pro-apoptotic protein BAX to PODS [25]. FALZ is a novel DNA-binding protein that can repress transcription, and it has been localized to a subset of amyloidcontaining plaques in the brains of AD patients [26]. These genes might explain the development of the neurologic features of RTT.
The expression levels of six genes (WDR42A, HNRNPA0, HIS1, RBM12, CYP7B1 and FOXC1) were decreased to below 25% of the control levels. Three genes (HNRPA0, HIS1, and FOXC1) have anti-apoptotic and proliferative functions [2127].
In the present study, TFBS was used to identify TFs in genes up-regulated by MeCP2. TFBS analysis is important for understanding the global effects of MeCP2 expression [28]. In silico analysis was carried out to match the promotor sequences of the identified target genes to their corresponding TFs in a large database [9]. Interactions between TFs and DNA are commonly described by position weight matrices, derived by aligning all known TFBSs and log transforming the observed number of each nucleotide at each position [29]. Match uses the position weight matrix table provided in the TRANSFAC database to calculate similarity scores to measure the quality of a match between the input sequences for comparison and the matrix in the database, where a value of 1.0 denotes an exact match [11]. It is important to choose appropriate cutoff values of similarity scores for optimal search results. In the present study, pre-calculated cutoff values minimizing false positives were chosen, which were derived from applying the search algorithm to the sequences of second exons where no biologically relevant TFBS would exist [11]. The results of the TFBS analysis were quite similar to those obtained for previously validated target genes, which confirms the reliability of the analysis result (Figure 3A) [30313233]. Importantly, the predicted TFs were related to tumorigenesis and RTT (Figure 3B).
Six transcription factors, including AP-1, AP-2, CREB, SP-1, USF, and NF-AT, have been implicated in an RTT model [223435]. The transcription repression domain of MeCP2 participates in transcriptional silencing via interaction with a corepressor complex containing mSin3A and HDAC1 [36]. Of the analyzed transcription factors, MyoD has been reported to bind to HDAC1, and it is functionally linked to the silencing of a muscle-specific regulatory gene prior to skeletal myogenesis in RTT [14]. C/EBPβ is associated with mSin3A/HDAC1 and is closely related to the pathogenesis of RTT [37].
Taken together, the results of the present study suggest that MeCP2 silencing affects apoptosis and cell proliferation (Figure 4). Many epigenetic factors including MeCP2 are known to be associated with tumorigenesis, suggesting that cancer is an epigenetic disease [38]. A limitation of this study is that the in silico TFBS analysis was not validated by another experiment, such as chromatin immunoprecipitation. Because this was an initial screening step for identifying target genes of MeCP2 utilizing cDNA microarray and computerized TFBS analysis, future studies must evaluate the significance of these targets and transcription factors in the pathogenesis of RTT and other diseases associated with MeCP2.
Figures and Tables
Figure 2
Expression levels of MeCP2 mRNA from HEK293 cells transfected with empty siRNA (control), with transfection reagent only (amine), or with MeCP2-siRNA with amine (siRNA-MeCP2). The level of MeCP2 mRNA in HEK293 cells treated with siMeCP2 was reduced by 60%.
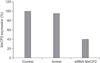
Figure 3
Potential transcription factor (TF) binding sites of regulated genes by MeCP2. (A) The shaded cells represent transcription factor binding sites. Cells with dark grey shading indicate TFs common to all of the 11 newly identified target genes of MeCP2; light grey colored cells indicate TFs common to 8–10 of the genes. (B) The predicted TFs were related to tumorigenesis and Rett syndrome. Fold change is reported as the log2 of Cy5 (siRNA-MeCP2)/Cy3 (control).

Figure 4
Regulation of proliferation and apoptosis influenced by MeCP2. Black boxes indicate newly identified candidate target genes; grey boxes represent predicted transcription factors.
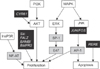
Table 4
Up-regulated and down-regulated genes with a greater than 4-fold change as a result of MePC2 silencing in HEK293 cells (HEK293siMeCP2)
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) and the Center for Women in Science, Engineering and Technology (WISET) Grant funded by the Ministry of Science, ICT & Future Planning of Korea (MSIP) under the Program for Returners into R&D. This work was also supported by the R&D program of MOTIE/KIAT (N0000697, Establishment of Infra Structure for Anti-aging Industry Support). The results of this work were included in the thesis for the PhD in science by Injoo Kim of the Graduate School of Pusan National University.
References
1. Rett A. Uber ein eigenartiges hirnatrophisches Syndrom bei Hyperammonamie im Kindesalter. Wien Med Wochenschr. 1966; 116(37):723–726.
2. Hoffbuhr K, Devaney JM, LaFleur B, Sirianni N, Scacheri C, Giron J, et al. MeCP2 mutations in children with and without the phenotype of Rett syndrome. Neurology. 2001; 56(11):1486–1495.

3. Imessaoudene B, Bonnefont JP, Royer G, Cormier-Daire V, Lyonnet S, Lyon G, et al. MECP2 mutation in nonfatal, non-progressive encephalopathy in a male. J Med Genet. 2001; 38(3):171–174.

4. Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003; 33:Suppl. 245–254.

5. Bernard D, Gil J, Dumont P, Rizzo S, Monte D, Quatannens B, et al. The methyl-CpG-binding protein MECP2 is required for prostate cancer cell growth. Oncogene. 2006; 25(9):1358–1366.

6. Billard LM, Magdinier F, Lenoir GM, Frappart L, Dante R. MeCP2 and MBD2 expression during normal and pathological growth of the human mammary gland. Oncogene. 2002; 21(17):2704–2712.

7. Darwanto A, Kitazawa R, Maeda S, Kitazawa S. MeCP2 and promoter methylation cooperatively regulate E-cadherin gene expression in colorectal carcinoma. Cancer Sci. 2003; 94(5):442–447.

8. Hite KC, Adams VH, Hansen JC. Recent advances in MeCP2 structure and function. Biochem Cell Biol. 2009; 87(1):219–227.
9. Wingender E. TRANSFAC, TRANSPATH and CYTOMER as starting points for an ontology of regulatory networks. In Silico Biol. 2004; 4(1):55–61.
10. Wingender E, Chen X, Fricke E, Geffers R, Hehl R, Liebich I, et al. The TRANSFAC system on gene expression regulation. Nucleic Acids Res. 2001; 29(1):281–283.

11. Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003; 31(13):3576–3579.

12. Pairo E, Maynou J, Marco S, Perera A. A subspace method for the detection of transcription factor binding sites. Bioinformatics. 2012; 28(10):1328–1335.

13. Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001; 98(9):5116–5121.

14. Hashimoto Y, Akiyama Y, Yuasa Y. Multiple-to-multiple relationships between microRNAs and target genes in gastric cancer. PLoS One. 2013; 8(5):e62589.

15. Hill DP, Adams N, Bada M, Batchelor C, Berardini TZ, Dietze H, et al. Dovetailing biology and chemistry: integrating the Gene Ontology with the ChEBI chemical ontology. BMC Genomics. 2013; 14:513.

16. Zhao LY, Zhang J, Guo B, Yang J, Han J, Zhao XG, et al. MECP2 promotes cell proliferation by activating ERK1/2 and inhibiting p38 activity in human hepatocellular carcinoma HEPG2 cells. Cell Mol Biol (Noisy-le-grand). 2013; Suppl 59. OL1876–OL1881.
17. Swerdlow RH. Mitochondrial DNA: related mitochondrial dysfunction in neurodegenerative diseases. Arch Pathol Lab Med. 2002; 126(3):271–280.

18. Nuber UA, Kriaucionis S, Roloff TC, Guy J, Selfridge J, Steinhoff C, et al. Up-regulation of glucocorticoid-regulated genes in a mouse model of Rett syndrome. Hum Mol Genet. 2005; 14(15):2247–2256.

19. Squillaro T, Alessio N, Cipollaro M, Renieri A, Giordano A, Galderisi U. Partial silencing of methyl cytosine protein binding 2 (MECP2) in mesenchymal stem cells induces senescence with an increase in damaged DNA. FASEB J. 2010; 24(5):1593–1603.

20. Rasti M, Arabsolghar R, Khatooni Z, Mostafavi-Pour Z. p53 Binds to estrogen receptor 1 promoter in human breast cancer cells. Pathol Oncol Res. 2012; 18(2):169–175.

21. Yang LH, Han Y, Li G, Xu HT, Jiang GY, Miao Y, et al. Axin gene methylation status correlates with radiosensitivity of lung cancer cells. BMC Cancer. 2013; 13:368.

22. Xu X, Jin H, Liu Y, Liu L, Wu Q, Guo Y, et al. The expression patterns and correlations of claudin-6, methy-CpG binding protein 2, DNA methyltransferase 1, histone deacetylase 1, acetyl-histone H3 and acetyl-histone H4 and their clinicopathological significance in breast invasive ductal carcinomas. Diagn Pathol. 2012; 7:33.

23. Bowser R, Reilly S. Expression of FAC1 in activated microglia during Alzheimer's disease. Neurosci Lett. 1998; 253(3):163–166.

24. Yanagisawa H, Bundo M, Miyashita T, Okamura-Oho Y, Tadokoro K, Tokunaga K, et al. Protein binding of a DRPLA family through arginine-glutamic aciddipeptide repeats is enhanced by extended polyglutamine. Hum Mol Genet. 2000; 9(9):1433–1442.

25. Joulie M, Miotto B, Defossez PA. Mammalian methylbinding proteins: what might they do? Bioessays. 2010; 32(12):1025–1032.

26. Chen Y, Luo J, Tian R, Sun H, Zou S. miR-373 negatively regulates methyl-CpG-binding domain protein 2 (MBD2) in hilar cholangiocarcinoma. Dig Dis Sci. 2011; 56(6):1693–1701.

27. Yan Z, Xiong Y, Xu W, Li M, Cheng Y, Chen F, et al. Identification of recurrence-related genes by integrating microRNA and gene expression profiling of gastric cancer. Int J Oncol. 2012; 41(6):2166–2174.

28. Jolly ER, Chin CS, Herskowitz I, Li H. Genome-wide identification of the regulatory targets of a transcription factor using biochemical characterization and computational genomic analysis. BMC Bioinformatics. 2005; 6:275.

29. Bernard B, Thorsson V, Rovira H, Shmulevich I. Increasing coverage of transcription factor position weight matrices through domain-level homology. PLoS One. 2012; 7(8):e42779.

30. Sapkota Y, Robson P, Lai R, Cass CE, Mackey JR, Damaraju S. A two-stage association study identifies methyl-CpG-binding domain protein 2 gene polymorphisms as candidates for breast cancer susceptibility. Eur J Hum Genet. 2012; 20(6):682–689.

31. Ping SY, Shen KH, Yu DS. Epigenetic regulation of vascular endothelial growth factor a dynamic expression in transitional cell carcinoma. Mol Carcinog. 2013; 52(7):568–579.

32. Samaco RC, Hogart A, LaSalle JM. Epigenetic overlap in autism-spectrum neurodevelopmental disorders: MECP2 deficiency causes reduced expression of UBE3A and GABRB3. Hum Mol Genet. 2005; 14(4):483–492.

33. Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent BDNF transcription, dendritic growth, and spine maturation. Neuron. 2006; 52(2):255–269.

34. Babbio F, Castiglioni I, Cassina C, Gariboldi MB, Pistore C, Magnani E, et al. Knock-down of methyl CpG-binding protein 2 (MeCP2) causes alterations in cell proliferation and nuclear lamins expression in mammalian cells. BMC Cell Biol. 2012; 13:19.

35. van Zuiden M, Geuze E, Willemen HL, Vermetten E, Maas M, Amarouchi K, et al. Glucocorticoid receptor pathway components predict posttraumatic stress disorder symptom development: a prospective study. Biol Psychiatry. 2012; 71(4):309–316.

36. Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998; 393(6683):386–389.

37. Wang JT, Wu TT, Bai L, Ding L, Hao M, Wang Y. Effect of folate in modulating the expression of DNA methyltransferase 1 and methyl-CpG-binding protein 2 in cervical cancer cell lines. Zhonghua Liu Xing Bing Xue Za Zhi. 2013; 34(2):173–177.




 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download