Abstract
Objectives
This paper will present an overview of the developmental effort in harmonizing clinical knowledge modeling using the Detailed Clinical Models (DCMs), and will explain how it can contribute to the preservation of Electronic Health Records (EHR) data.
Methods
Clinical knowledge modeling is vital for the management and preservation of EHR and data. Such modeling provides common data elements and terminology binding with the intention of capturing and managing clinical information over time and location independent from technology. Any EHR data exchange without an agreed clinical knowledge modeling will potentially result in loss of information.
Results
Many attempts exist from the past to model clinical knowledge for the benefits of semantic interoperability using standardized data representation and common terminologies. The objective of each project is similar with respect to consistent representation of clinical data, using standardized terminologies, and an overall logical approach. However, the conceptual, logical, and the technical expressions are quite different in one clinical knowledge modeling approach versus another. There currently are synergies under the Clinical Information Modeling Initiative (CIMI) in order to create a harmonized reference model for clinical knowledge models.
For many healthcare professionals there is no greater contrast between their main professional value of human centered care for a person with an illness, and the digitized numbers crunched by computers. This contradiction is very fundamental because healthcare professionals' baseline values imply individualization of tailored care. In contrast, automation is still not beyond the stage where the Von Neumann principle requires any computer activity to be transformed into sets of binary values, manageable through the funnel of the computer processor, albeit nowadays in parallel. This contradiction materializes often in a perceived or real resistance of clinicians towards computer use in healthcare in general, and Electronic Health Records (EHR) in particular.
Authors consider two main developments that might lead to a change in the future. Healthcare professionals realize they cannot afford information processing by hand anymore, urging a systematic approach to information management. The challenge for healthcare can be summarized as "offer better quality of care with less money and decreasing numbers of professionals to a growing population of elderly people with increasing numbers of chronic diseases." The second development is that software can be realized in increasingly shorter development cycles, based on model driven approaches, user friendly interfaces, parallel processing, and other improvements in the world of information and communication technology. Their impact is that user requirements can be met much easier today than in the time each command had to be entered after the prompt.
One particularly interesting novelty in this space is the development and deployment of so called Detailed Clinical Models or DCMs [1,2,3]. ISO/TS 13972 defines a DCM as "an information model designed to express one or more clinical concept(s) and their context in a standardized and reusable manner, specifying the requirements for clinical information as a discrete set of logical clinical data elements [2]." So a model, hence reduction of reality, with clinical knowledge, so a focus on healthcare, and it specifies concepts and data elements, so meaning is put into computable environments. The purpose of this paper is to give further explanation about the goals, requirements, position and relevance of DCMs and how they can contribute to the long-term preservation of healthcare data.
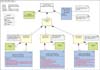
In the early nineties, a group of smart scientists in the area of health informatics invented an approach to the development of EHRs which was called two level modeling [4]. In this approach, the generic functions of record systems, such as entering, storing, presenting, managing, communicating, and adding generic data, such as patient name and ID, location, time, professional (e.g., through the login), where separated out from the clinical details. And, these clinical details can be millions, and only need specific descriptions to distinguish these from each other. Figure 1 illustrates the distinction [5].
Following this approach several initiatives emerged in which the clinical knowledge was specified, so that computers can operate on this and generate meaningful information. These various approaches have been reviewed [3]. Different initiatives to create collections of clinical knowledge models include the OpenEHR archetypes [6], Intermountain Healthcare clinical element models [1], the Netherlands care information models [7], and South Korea clinical contents models [8], among others. Following the original separation suggested by Rector et al. [4], these projects do lead to similar descriptions of health-related data and in some instances their context.
However, there are differences as well [3]. Some do not include the medical background knowledge, some do apply standardized terminologies and codes, others do not, or use internal code mechanisms. Also, differences exist in the logical formats and technical representations applied. In particular the following specification methods are used: Archetype Definition Language (ADL), Unified Modeling Language (UML) and/or eXtensible Markup Language (XML). There is also differs in the use of a reference model or not. A reference model imposes specific characteristics to the clinical models, to let these fit in a specific system or implementation specification. Given the gap mentioned in the introduction, we also see efforts to create tools to communicate with professionals. Due to the quite significant differences, these models cannot be used from one project to the other. Partly this is because of the ever-existing cultural differences and realm specifics. However, if we move beyond that, we see ADL dialects (e.g., ISO 13606 versus OpenEHR based), UML variants (e.g., proper specifications from the Object Management Group versus the Health Level 7 [HL7] colorful dialect), and in the XML space, each project has their own tag names and build up. So, despite the common ground: i.e., the same medical knowledge, the same data elements, the same codes from standardized terminologies, these efforts do not deliver full semantically interoperable specifications, due to requiring the systems to adhere to specific reference models and computational formats.
Time for a change! With the DCM approach the benefits of the two level modeling are respected and the enormous efforts that went into the clinical modeling efforts are also acknowledged. The idea dates back to 2007 where during a workshop, an overarching approach called Detailed Clinical Modeling emerged, using a term from Huff et al. [1]. This approach takes the commonalties of the various specifications to keep the conceptual descriptions and the logical models of data elements, code binding, relationships, data type specifications and such [9]. However, it does not go to the level of physical specification, and even, the logical model can easily be converted to reference model based approaches as Health Level 7 Reference Information Model (HL7 RIM) or OpenEHR or reference models.
Due to the interoperability issues among these clinical modeling approaches and the investments already made, there currently is an approach trying to harmonize the existing work. This is carried out by the Clinical Information Modeling Initiative (CIMI). Their goal is to create a harmonized reference model for clinical knowledge models using both the ADL and UML formalisms. From these baseline formats, any kind of technical artifact can potentially be derived. CIMI also works on an EHR clinical model repository as open content. However, in practice, with all good intentions, CIMI has created yet another reference model and yet another dialect of ADL. Nevertheless, other initiatives seem to start picking these harmonization results up.
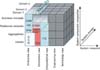
Modern development techniques apply often the Model Driven Architecture (MDA). In this, models are important, and reside at the logical level mostly. DCMs have a place in MDA. This can best be explained using the Generic Component Model (GCM) [10]. This cubical model positions DCMs in healthcare architectures, using a three-dimensional space. GCM characterizes any system by three axes: domain, system components, and system development (Figure 2).
At the system axis (x-axis), the Reference Model of Open Distributed Processing (RM-ODP) serves as a coordinating framework. This framework comprises with five components namely the enterprise viewpoint, information viewpoint, computational viewpoint, engineering viewpoint, and finally technical viewpoint. The RM-ODP positions DCMs in the enterprise, information and the computation space (e.g., for detailed computational specifications, such as calculation of total scores on data and such).
The second axis (y-axis) specifies the system development approach of the MDA. MDA separates the healthcare business and the application logic of EHRs from the specific implementation technology [11]. According to Blobel [10], this MDA depends on standards, traceability, and explicit relationships between system components. And at the lowest level of clinical detail, that is what DCMs provide: consistency, traceability, and reusability, while covering the conceptual and logical levels. DCMs fit into larger logical models, such as reference (information) models.
At the domain axis (z-axis), the different healthcare domains, such as clinical specialties, are depicted. It is represented from the business at the top, to the fine grained data elements on the bottom. The latter specified in DCMs, reusable from domain to domain.
On the physical level, the DCMs based health data from EHRs must be storable and remain available for many years and in numerous technologies. Looking at applicable technologies for data preservation is relevant for clinical modeling exercises.
File format conversion engines that are constrained to one data type and in-house software base are available [12]. For example, FileFormat.Info (http://www.fileformat.info) includes file format conversion tools for images only based on Java Advanced Imaging libraries (javax.imageio.* and javax.media.jai.*). There exist a few file format conversion services that support only certain conversion types (e.g., http://www.ps2pdf.com 1 conversion type; http://media-convert.com about 20 multi-media formats; http://www.zamzar.com selected conversions of document, image, music, video, and couple of CAD formats). The main drawback of the existing conversion systems is that they are not extensible (limited by the availability of specific libraries).
In order to design an extensible file format conversion system based on utilizing third party software several problems have to be addressed [12]. First, the problem of automated execution of the software, most GUI based, without having access to an application programming interface. AutoHotkey (http://www.autohotkey.com) scripting is a viable option for the Windows operating system and the current Polyglot implementation is based on it. Second, the problem of distributed computational resources has been approached in the past by the Grid community, TeraGrid (https://www.xsede.org/tg-archives) and Globus Toolkit (http://toolkit.globus.org/toolkit/), for building computational grids, and the design of workflow middleware that would manage the execution, such as DAGMan, CCA (http://www.cca-forum.org) or Taverna (http://www.taverna.org.uk/), among others.
Due to the heterogeneity of computational hardware, this problem also requires considerations about options for parallel processing [12], for instance, the use of 1) a message passing interface is designed for the coordination of a program running as multiple processes in a distributed memory environment by using passing control messages; 2) open multi-processing is intended for shared memory machines. It uses a multithreading approach where the master threads fork any number of slave threads; 3) the map reduce parallel programming paradigm for commodity clusters which allows programmers write simple Map and Reduce functions, which are then automatically parallelized without requiring the programmers to code the details and communications of parallel processes; and 4) novel architectures FPGAs, GPUs, multiple CPUs. Unfortunately, none of the existing grid solutions are an option when utilizing 3rd party binaries compiled for specific hardware on one machine.
Workflow solutions could potentially orchestrate calling computational resources based on a conversion sequences, however most do not robustly deal with solely GUI based software and also tasks specific needs must be considered, such as clustering the conversion execution sequence into segments that do not require data movement, and then managing and monitoring entire conversion executions [12].
Such technologies would contribute to the preservation of data in EHRs and their use in various systems, using various technical formats and representations of the same clinical data based on DCMs.
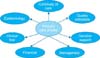
Healthcare has several different purposes for use of clinical data, such as clinical care, continuity of care, quality indicators, decision support, management, billing information, clinical trials, and epidemiological studies among others [9]. This is illustrated in Figure 3. Each purpose requires analysis of the requirements, in particular data granularity, validity, relevance, preciseness, and reliability. Each data use implies a specific set of attributes and constraints for the data entry, storage, processing, presentation, communication, selection, and aggregation. However, important is also that each data reuse from an EHR poses validity and reliability questions. For example, are there biases, confounders, and other factors to be taken into account? And finally, each data use has its own expectations for data preservation. The assumption is that clinical data are recorded into an EHR system during the primary care process, and that DCMs are applied to guide the data entry. For each purpose the DCMs serve as the semantic baseline. For instance, for continuity of care, the DCMs will be transformed into a HL7 v3 XML format. Hence they serve to define the message or document payload. Here the DCMs prove their value. It might be that all data according to the DCM are stored in the EHR, but not all data need to be exchanged. At the message/document definition a selection of the data can be made. However, each data element exchanged will still have all of the standardized characteristics according the DCM.
For all other purposes, similar mechanisms will apply. All data in EHR are available, process of selection might lead to minimizing the amount of data required. And then the aggregation process will add some features. For instance for a quality indicator, the denominator can be a data element standardized in a DCM, while the aggregation process requires a nominator as numeric value derived from the number of occurrences of the denominator, e.g., 'pressure ulcer risk present' as denominator specified as resulting data from an risk assessment, and the occurrence of 80 in a patient population of 320 would reveal an incidence rate of 25%. The data element handled comes from the DCM. The calculations are then based on scientific methods for incidence rates, and some policy might determine that the percentage of patients at risk for pressure ulcer might be a good quality indicator. However, in actual care, this indicator might need a second standard, DCM based, data element to be present and handled similarly, e.g., "patient receives preventive measures for pressure ulcer." If 80 of the 320 patients would get preventive care, this would be 25%, and a perfect match to the risk, and hence perfect care. For most goals, a selection of data elements according DCMs will be used, but still keep its standard features.
In ISO/TS 13972 the characteristics for DCMs are specified [2]. These have been summarized by Goossen [9] and are presented in Table 1 below. This implies that a proper DCM has a linkage to the medical knowledge about the concept that is modeled. And for each model, the data elements must be specified extensionally, with all core parts included. Further, lack of metadata, such as authorship, versioning and endorsement, is seen as a risk to patient safety, and should therefore be added to each DCM. Reader is referred to the TS for the complete listing of criteria for DCMs.
Comparing existing formats for the same medical concepts reveals the residue that is the core clinical content specified in DCMs. Goossen and Goossen-Baremans [13] carried out an analysis of clinical concepts in the format of archetype, HL7 v3 model, and DCM in UML format. Although that approach is using a specific bottom up analysis, and looked at data types and code bindings, it also showed the overall models. In this paper, we present a similar analysis on logical model level of the Glasgow Coma Scale (GCS) [14]. The GCS is used to determine the level of consciousness of patients after trauma, with stroke, or for other head injuries [14]. The GCS consists of three categories of data, representing eye opening, best motor response and best verbal response that are summed up into a total score. The GCS is scored by documenting the number representing the best response for each category that could be observed with the patient. Table 2 specifies the conceptual knowledge about the GCS.
The different expressions of the GCS in the different modeling approaches are illustrated below. For the ADL and XML, the basic parts showing the semantics are presented. Many technical ADL and XML specifications have been left out for easy reading. First the GCS core as an archetype version will be depicted (Figure 4).
Finally, the UML representation of a DCM is shown (Figure 7) and a preliminary representation using the recent HL7 Fast Healthcare Interoperability Resources (FHIR) approach (Figure 8). Note that a DCM can be expressed in any logical modeling method. UML is just for the illustration of the commonalties and differences. An example DCM is represented in Figure 7, using UML.
Finally, HL7 is currently investing in the new format for data exchange, FHIR [15]. The power to preserve investments in DCMs is illustrated best that in a very short time the FHIR resource exporter could be added to the used modeling tool, and export of FHIR resource format is available. Figure 8 illustrates a fragment of the FHIR resource, showing the four core data elements of the GCS. Note that to keep this readable, the XML parts that define the GCS and its values in FHIR have been left out of the example, and none core parts have also been removed. FHIR allows both the Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT) coding [16] and the Logical Observation Identifiers Names and Codes (LOINC) coding [17] to be present in the definition. Again, we only show here LOINC codes as example.
What these examples illustrate clearly is that the medical knowledge is the same; each model has the core components of the GCS expressed. And that must be so to ensure semantic interoperability. However, due to the technological choices, it is obvious that the technical specification part for each implementation specification differs. The art of modeling requires that we try every attempt to move from the technological approach to the clinician, and this can be done through the logical modeling of the conceptual content. In other words, let the technicians and modelers deal with the intricacies of modeling, do not bother doctors and nurses with it. But offer tooling that allow to do this consistently so that every implementation format for the computational level can use it adequately.
These results indicate that it is feasible to compare and reuse information models for single or combined clinical data elements and for assessment scales from one implementation approach to the other [13,18]. When the specific limitations of each approach are taken into account and a precise analysis of each data-item is carried out, it is possible to reveal the semantics of the different models, abstract from that and transform it into another logical model. In particular, the HL7 template approach and the ISO/CEN 13606 and OpenEHR archetypes reveal more commonalties than differences. Semantics are about the interaction between the medical knowledge represented in the clinical concepts, the information model representing it in technology, and the terminology model revealing its semantics [13]. The presented models do have a generic and equivalent structure where these concepts fit. The structures include for HL7 v3 the Clinical Statement Pattern, and for the 13606 and OpenEHR archetypes the Entry level. Both structures allow 1 - n data elements to be represented and linked together. The DCM example in UML applies a full class diagram in which the concept is modeled; each data element is represented in a class. According to Goossen and Goossen-Baremans [13], the best level of comparing HL7 v3 and OpenEHR is at the Clinical Statement Pattern versus Entry level, respectively, where both express a single clinical relevant data element. However, concepts do partly get their meaning from the structure they are embedded in. Hence this bottom up approach will lead to 100% basic semantics equivalence for data elements, but it will never lead to a 100% comparability of the much more abstract reference models.
However, it is possible to extract data from storage in one formalism and represent it into another formalism. And for the preservation of data that is very important.
The proper understanding of data in healthcare, and their proper representation which is necessary to bridge the gap between human centered caring for individuals and the zero and one conversion of computer processing of data, does require that the two level modeling is optimal. In this approach, basis system functions and clinical content specification are separated. The clinical models fit in different healthcare architectures and approaches, but are in each approach on the maximum (some say optimal) level of granularity.
This MDA, with its different axis as illustrated with the GCM can be done through several examples of clinical model specification that have in common the representation of knowledge, data, and semantics, but differ due to technological choices. The illustration of the GCS reveals that the core medical content is kept in the logical part of each modeling approach, facilitating reuse from one format to the other and allowing interoperability become true in some not to far away near future. For the preservation of clinical data this is important. Because besides the technical storage mechanisms, also the computational formalisms might cause difficulties on the long run. This paper covered the conceptual example of one scale, the GCS, and its logical representation in various formats. Fragments of the computational specifications have been presented, albeit incomplete. One important step which must be dealt with for each implementation technical specification is to test for the model and syntax correctness. For instance through XML schema and similar validations, or for the content through XML schematron.
The ISO/TS 13972 on DCM specifies the key characteristics of properly modeled clinical content, and its application to modeling will facilitate the reuse of models that are created elsewhere using a different approach. The work of CIMI is currently undertaking a first international effort to share each others models, once they have been abstracted and specified in a formalism that all can apply. This approach will contribute to the preservation of clinical data over time, location, and for various purposes, provided that the technical implementations will still be adequately tested.
Figures and Tables
Figure 5
Glasgow Coma Scale represented in Health Level 7 version 3 Reference Information Model classes.

Figure 6
Glasgow Coma Scale in Health Level 7 version 3 eXtensible Markup Language (XML) formalism (derived from the classes in Figure 5).

Figure 7
Detailed Clinical Models (DCMs) in Unified Modeling Language of the Glasgow Coma Scale (2014 revised but unpublished version).
Acknowledgments
Author wishes to acknowledge the contribution to the outline of the paper and substantive content of paragraph 3 of Mr. Wo Chang of the National Institute of Standards and Technology, Gaithersburg, MD, USA. Parts of this paper have been published before in Dutch. This work was undertaken as volunteer work for the CODATA conference in Taiwan in 2012.
References
1. Huff SM, Rocha RA, Coyle JF, Narus SP. Integrating detailed clinical models into application development tools. Stud Health Technol Inform. 2004; 107(Pt 2):1058–1062.
2. International Organization for Standardization. Health informatics: detailed clinical models, characteristics and processes. Geneva, Switzerland: International Organization for Standardization;2014. (ISO/PRF TS 13972).
3. Goossen W, Goossen-Baremans A, van der Zel M. Detailed clinical models: a review. Healthc Inform Res. 2010; 16(4):201–214.

4. Rector AL, Nowlan WA, Kay S, Goble CA, Howkins TJ. A framework for modeling the electronic medical record. Methods Inf Med. 1993; 32:109–119.
5. Goossen W. Detailed clinical models: kennis en semantiek weergeven met UML en XML. <!Element>. 2011; 17(1):11–16.
6. Beale T. Archetypes and the EHR. Stud Health Technol Inform. 2003; 96:238–244.
7. van der Kooij J, Goossen WT, Goossen-Baremans AT, Plaisier N. Evaluation of documents that integrate knowledge, terminology and information models. Stud Health Technol Inform. 2006; 122:519–522.
8. Center for Interoperable EHR (CiEHR). Clinical Contents Manager [Internet]. Seoul, Korea: CiEHR;c2012. cited at 2012 Oct 16. Available from: http://www.clinicalcontentsmodel.org.
9. Goossen W. Representing knowledge, data and concepts for EHRS using DCM. Stud Health Technol Inform. 2011; 169:774–778.
10. Blobel B. Architectural approach to eHealth for enabling paradigm changes in health. Methods Inf Med. 2010; 49(2):123–134.

11. Object Management Group. Model Driven Architecture (MDA). Needham (MA): Object Management Group;c2014. cited at 2014 Jul 1. Available from: http://www.omg.org/mda.
12. Goossen W, Chang W. Clinical knowledge modeling using Detailed Clinical Models. In : Proceedings of the 23rd International CODATA Conference; 2012 Oct 27-31; Taipei, Taiwan.
13. Goossen WT, Goossen-Baremans A. Bridging the HL7 template: 13606 archetype gap with detailed clinical models. Stud Health Technol Inform. 2010; 160(Pt 2):932–936.
14. Teasdale G, Jennett B. Assessment of coma and impaired consciousness: a practical scale. Lancet. 1974; 2(7872):81–84.
15. Health Level Seven International. Fast Healthcare Interoperability Resources (FHIR) [Internet]. Ann Arbor (MI): Health Level Seven International;c2014. cited at 2014 Jul 1. Available from: http://wiki.hl7.org/index.php?title=FHIR.
16. International Health Terminology Standards Development Organisation. SNOMED CT [Internet]. Copenhagen, Denmark: International Health Terminology Standards Development Organisation;c2014. cited at 2014 Jul 1. Available from: http://www.ihtsdo.org/snomed-ct/.
17. Regenstrief Institute Inc. LOINC [Internet]. Indianapolis (IN): Regenstrief Institute Inc.;c2014. cited at 2014 Jul 1. Available from: http://loinc.org/.
18. Cuggia M, Bayat S, Rossille D, Poulain P, Pladys P, Robert H, et al. Comparing the APGAR score representation in HL7 and OpenEHR formalisms. Stud Health Technol Inform. 2009; 150:250–254.




 PDF
PDF ePub
ePub Citation
Citation Print
Print







 XML Download
XML Download