Abstract
A multiplex PCR method has been developed to classify extended spectrum β-lactamase (ESBL) and plasmid-mediated AmpC β-lactamase (PABL). This method consists of the use of two four-multiplex PCRs for the detection of TEM, OXA, SHV, CTX-M, CMY, and DHA type β-lactamases. We have compared findings from the use of conventional detection methods with that of this newly developed typing method. In testing for 73 ESBL-producing and PABL-producing isolates, 100% of the isolates were correctly identified as previously characterized types and, 44 types of β-lactamases were additionally identified from 33 isolates. This assay not only reduces the time for classification but also increases the accuracy for detection.
Extended spectrum β-lactamases (ESBLs) and plasmid-mediated AmpC β-lactamases (PABLs) hydrolyze a variety of cephalosporins (1-4). Recently, resistance to cephalosporins has become widespread throughout the world, and numerous types of ESBLs and PABLs have been detected in various bacterial organisms (5). Detection and classification of ESBLs and PABLs are important in makingfor clinical decisions regardingfor appropriate therapy and infection control. However, detection and classification are time-consuming and complicated processes, and some types of ESBLs and PABLs are frequently not detected during the detection process (1).
In this study, we described a multiplex PCR method to classify common ESBLs and PABLs and we evaluated the efficiency and accuracy of this method as compared to the conventional method.
Seventy-three (3 Enterobacter cloacae, 12 Escherichia coli, 21 Shigella sonnei, and 37 Klebsiella pneumoniae isolates) ESBL-producing and PABL-producing isolates obtained from the Korea Centers for Disease Control and Seoul National University Children's Hospital collection were used in this study (6, 7). All isolates were previously typed for β-lactamase according to usinge the of a conventional method.
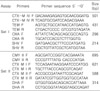
Suitable primers for two multiplex PCR reactions, each targeting four different regions, were designed using Primer Express v2.0 (Applied Biosystems, Foster City, CA, USA), which and were synthesized by Bioneer co. (Daejeon, Korea). The first multiplex assay (named Set I) was designed to detect TEM, SHV, CTX-M IV group (8-10), and OXA β-lactamase encoding genes, and the second assay (named Set II) was designedto detect CTX-M I group, CTX-M II group, CMY II, and DHA encoding genes (http://www.lahey.org/studies/webt.asp) (Table 1). Both PCR reactions were performed under identical conditions and optimal results were obtained using TaKaRa Ex Taq polymerase (Takara, Tokyo, Japan). Reactions were performed in a final volume of 25 µL containing 5 µL of template DNA, 1× reaction buffer, 0.2 mM of each deoxynucleoside triphosphate, 20 pM of each primer, and 3.5 units of Taq polymerase. Both assays used identical cycling conditions. Reactions were performed in a Peltier Thermal Cycler (MJ Research, Waltham, MA, USA) under the following conditions: denaturation at 94℃ for 5 min followed by 30 cycles of 94℃ for 1 min, 61℃ for 1 min and 72℃ for 1 min, and a final extension of 72℃ for 5 min. After PCR amplification, 3 µL of each reaction was separated by electrophoresis 2% agarose gel (NuSieve 3:1, FMC Bioproducts, Rockland, NY, USA). Both assay products were electrophoresed for 30 min at 100 V in 0.5× TBE buffer. DNA was stained with ethidium bromide (1 µg/mL) and the gels were imaged under UV light. PCR amplicon sizeswere calculated by a comparingson with to molecular weight size markers.
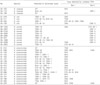
We optimized the conditions of both multiplex PCR reactions to ensure that each amplicon was of the correct size and that a sufficient quantity of product was generated to permit easy detection after gel electrophoresis (Fig. 1). All of the Set I and Set II assays produced single or multiplexed products of the predicted sizes (Fig. 1). After optimization, the assay method was tested on the 73 isolates that produced ESBLs, PABLs or both, where the isolates had already been confirmed by the use of conventional methods. Of the previously known ESBLs and PABLs, those produced by each of the 73 tested isolates were successfully identified with the use of the multiplex assays. We identified additional bla genes from 33 isolates (3 E. cloacae, 4 E. coli, 9 S. sonnei, and 17 K. pneumoniae) that had not been previously identified with the use of conventional methods (Table 2). The additionally identified β-lactamase-encoding genes in the Set I assay were 14 SHV, 8 OXA, 6 TEM and 4 CTX-M-IV types. These were identified from 2 E. cloacae, 4 E. coli, 3 S. sonnei, and 17 K. pneumoniae isolates. In four of the K. pneumoniae isolates, two additional bla genes were identified from each isolate and three additional bla genes were identified from E. coli 02-542. Because the SHV-1-like enzymes are ubiquitous in K. pneumoniae, detections of these enzymes in K. pneumoniae are expected (11). For the Set II assay, additional types of bla genes were identified from 12 isolates: 1 E. cloacae, 1 E. coli, 8 Shigella spp., and 2 K. pneumoniae isolates. The additional types identified by the Set II assay were 1 CTX-M-1 group, 9 CMY II, and 2 DHA types. The CTX-M-I encoding gene amplified from S. sonnei 99-1505 was further identified as blaCTX-M-15 by DNA sequencing, and this was the first blaCTX-M-15 subtype identified from Shigella spp. isolated in Korea.
Classification of ESBL and PABL using phenotypic methods is very difficult since, as many bacterial pathogens are resistant to third-generation cephalosporins, produce two or more ESBLs and/or PABLs, or have overlapping phenotypes. Therefore, isoelectric focusing (IEF) and type-specific PCR have been employed to classify ESBLs and PABLs. IEF is usually performed first, and the type deduced from the use of IEF is confirmed by type-specific PCR. However, as many of the isoelectric points of the various β-lactamases overlap (1, 8), IEF has only limited detection ability. Therefore, the combination of IEF and type-specific PCR can prevent detection of some ESBLs and PABLs that are present in extended spectrum cephalosporin-resistant bacteria, and furthermore, this combination method is time-consuming.
The multiplex PCR assay developed in this study not only simplifies the detection of ESBLs and PABLs, but also increases the detection ability. This assay takes only 4-5 hours to perform and with the use of this assay, we identified β-lactamases from 33 isolates among the 73 ESBL-producing or PABL-producing isolates where the ESBL and PABL types had already been confirmed by use of conventional methods. Seven additional types were identified, and as there was no overemphasis of one or a few types. We believe that we have developed a fast and accurate assay that will be useful in infectious disease control and prevention.
Figures and Tables
 | Figure 1Amplification profiles of each primer set for the Set I and Set II multiplex assays are shown. Set I assay products are shown in the first five lanes with the primer pairs indicated above each lane. (Lane 6 was empty.) Set II assay products are shown in the next five lanes with the primer pairs indicated above each lane. The sizes of the DNA standards are indicated (in base pairs) to the left of the gel image. Amplicon sizes for the Set I reactions are indicated on the left side of the gel, while amplicon sizes for the Set IIreactions are indicated on the right side of the gel. |
Acknowledgements
The authors wish to thank Dr. R. Ben-Ami and Dr. Shiri Navon-Venezia for providing CTX-M-2 DNA.
This research was supported by a grant by Korea Centers for Disease Control and Prevention.
References
1. Bradford PA. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001. 14:933–951.

2. Bush K. New beta-lactamases in gram-negative bacteria: diversity and impact on the selection of antimicrobial therapy. New beta-lactamases in gram-negative bacteria: Diversity and impact on the selection of antimicrobial therapy. Clin Infect Dis. 2001. 32:1085–1089.
3. Pai H, Kang CI, Byeon JH, Lee KD, Park WB, Kim HB, Kim EC, Oh MD, Choe KW, Pai H, Kang C, Byeon J, Lee K, Park B, Kim H, Kim E, Oh M, Choe K. Epidemiology and clinical features of bloodstream infections caused by AmpC-type-beta-lactamases-producing Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004. 48:3720–3728.

4. Pai H, Hong JY, Byeon JH, Kim YK, Lee HJ. High prevalence of extended-spectrum beta-lactamases-producing strains among blood isolates of Enterobacter spp. collected in a tertiary hospital during an 8-year period and their antimicrobial susceptibility patterns. Antimicrob Agents Chemother. 2004. 48:3159–3161.

5. Shah AA, Hasan F, Ahmed S, Hameed A. Characteristics, epidemiology and clinical importance of emerging strains of Gram-negative bacilli producing extended-spectrum beta-lactamases. Res Microbiol. 2004. 155:409–421.

6. Lee J, Pai H, Kim YK, Kim NH, Eun BW, Kang HJ, Park KH, Choi EH, Shin HY, Kim EC, Lee HJ, Ahn HS. Control of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in a children's hospital by changing antimicrobial agent usage policy. J Antimicrob Chemother. 2007. 60:629–637.

7. Kim S, Kim J, Kang Y, Park Y, Lee B. Occurrence of extended-spectrum-beta-lactamases in members of the genus Shigella in the Republic of Korea. J Clin Microbiol. 2004. 42:5264–5269.

8. Bonnet R. Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother. 2004. 48:1–14.

9. Pitout JD, Hossain A, Hanson ND. Phenotypic and molecular detection of CTX-M-beta-lactamases produced by Escherichia coli and Klebsiella spp. J Clin Microbiol. 2004. 42:5715–5721.

10. Amino acid sequences for TEM, SHV and OXA extended-spectrum and inhibitor-resistant β-lactamases. Lahey Clinic. Available from: URL:
http://www.lahey.org/studies/webt.asp.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download