Abstract
Resistance of falciparum malaria to antimalarial agents is prevalent in many areas, whereas chloroquine-resistant vivax malaria has been reported mainly around New Guinea since 1989. Concomitant with the spread of chloroquine-resistant P. vivax and increase in number of international travelers, imported cases of chloroquine-resistant vivax malaria in travelers returning from these areas has been reported. We experienced a case of chloroquine resistance P. vivax infection imported from Mangole Island, Indonesia. Its origin is confirmed not to be indigenous by the gene encoding analysis for the polymorphic region of apical membrane antigen-1 in P. vivax. Gene sequencing of the P. vivax mdr1 gene revealed only one substitution located at the codon 1076 (F1076L). The case was managed with oral quinidine with successful outcomes.
Malaria is one of the most threatening infections worldwide, and results in more than 150 million deaths annually. Vivax malaria is not normally associated with mortality, but does cause morbidity of long duration, relapses, and is also associated with the delivery of low birth-weight infants. Since the development of chloroquine as a therapeutic or prophylactic agent against malaria, malaria-associated mortality and morbidity have declined significantly. However, in the 40 years since chloroquine-resistant falciparum malaria was first discovered in Colombia in 1961, chloroquine-resistant falciparum malaria has spread to almost every region in which malaria is detected. More recently, P. vivax (1) and P. malariae (2) have also shown evidence of some chloroquine resistance. The rates of chloroquine-resistant P. vivax in Papua New Guinea and Indonesia are approximately 50%, and sporadic cases of chloroquine- resistant P. vivax have also been detected in other areas, including Malaysia, Vietnam, Myanmar, and South American countries (3). In this report, we describe a case of chloroquine-resistant P. vivax infection in a person returning from Indonesia, which was successfully managed by the administration of oral quinidine.
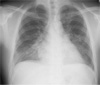
A Fig.1. Chest radiography on day-5 shows bilateral pulmonary infiltrates. 24-year old, 64 kg-weighted sailor was hospitalized because of fever of 12 days' duration. He had been residing in Incheon, Korea and never visited the malaria endemic areas in Korea in the recent 3 years. 25 days ago, he visited the Pulau Mangole (Mangole Island), Indonesia and stayed there from January 23 to February 2, 2004 without antimalarial prophylaxis. During the return to Korea by sea, he felt febrile sensation and weakness sine from February 4. With only symptomatic treatment, his fever became worse resulting in the admission on February 16, the day of his arrival in Korea. On admission, the body temperature was 39.2°. Pale conjunctivae and hepatosplenomegaly were noted. Hemoglobin level was 11.6 g/dL, white blood cell count 4,200/ mm3, and platelet count 79,000/mm3. Serum bilirubin level was 1.4 mg/dL, aspartate aminotransferase 73 IU/L, and alanine aminotransferase 118 IU/L. Peripheral blood smear showed trophozoites of P. vivax at the concentration of 1,380/mm3. A total of 2,000 mg of hydroxychloroquine (800 mg on Day 1 and three doses of 400 mg, 6 hours, 24 hours, and 48 hours later, respectively) did not decreased his fever. Hydroxychloroquine was administered by nurse and there was neither emesis nor diarrhea. On day 5, body temperature was 40.4°, parasitemia rose to 6,318/ mm3, and hemoglobin level dropped to 8.4 g/dL. New onset of cough developed, which was later confirmed as the result of pulmonary edema in chest PA (Fig. 1). The absence of concomitant P. falciparum was confirmed by polymerase chain reaction and by the P. falciparum histidine-rich protein-2 (PfHRP-2)-based assay (ICT Malaria Pf/Pv assay) (ICT Diagnostics, Sydney, Australia). Unfortunately the blood level of chloroquine was not measured regretfully. On the impression of chloroquine-resistant P. vivax or possible co-existing unrecognized falciparum malaria, oral quinidine sulfate (600 mg three times a day for 7 days) was administered resulting in abatement of fever over 2 days. Anemia, leukopenia, thrombocytopenia and pulmonary edema were improved and peripheral blood smear taken 5 days later showed no malaria parasite (Table. 1). After his fever became absent, a course of high-dose primaquine (primaquine base 30 mg/day) was administered for 14 days. Blood smear examined on Day 23 showed no parasitemia.
Plasmodium vivax genomic DNAs were extracted from the patient's blood by using a QIAamp DNA Blood Kit (Qiagen, Valencia, USA) according to the manufacturer's instruction. In order to confirm the diagnosis of malaria and to differentiate between P. falciparum and P. vivax infections, single-step polymerase chain reactions (PCRs) were performed as previously described(4). The PCRs showed only P. vivax.
Polymorphic region of the gene encoding for apical membrane antigen-1 (AMA-1) were analyzed to evaluate population diversity(5). The polymorphic region of the AMA-1 gene was amplified by PCR, and the amplified product was cloned and sequenced as previously described(6). Analysis of the AMA-1 gene sequence revealed that the isolate was classified into the genotype which had previously been identified in Indonesia and other Southeast Asian areas (Plasmodium vivax strain Salvador I).
To detect and analyze mutations in pvmdr1 gene, polymerase chain reaction was done with 100 ng of P. vivax genomic DNA, using specific oligonucleotides for pvmdr1 as described previously(7). The forward primer was 5'-ATGAAAAAGGATCAAAGGCAAC-3' and reverse primer was 5'-CTACTTAGCCAGCTTGACGT AC-3'. The amplified products were gel-purified, ligated into the pCR2.1 vector, and transformed into competent Escherichia coli Top10 cells, using a TOPO TA Cloning Kit (Invitrogen, Carlsbad, USA). Sequencing reactions were conducted with a BigDye Terminator Cycle Sequencing Ready Reaction Kit in an ABI 377 automatic DNA sequencer (Applied Biosystems, Foster City, USA). To verify the relevant sequences, we analyzed multiple plasmid clones containing each insert. Nucleotide and deduced amino acid sequences were analyzed with the SeqEd.V1.0.3 program, and the CLUSTAL program provided in the Megalign software, a multiple-alignment program of the DNASTAR package (DNASTAR, Madison, USA). Analyses of pvmdr1 gene showed that the F1076L substitution was present in the P. vivax isolate evaluated (Fig. 2).
In Korea, relapses after standard therapy for imported vivax malaria are attributed to the presence of primaquine tolerance(8, 9), and the recurrence or persistence of vivax malaria after chloroquine therapy has never been reported. Although one report describes a case of recurrent vivax malaria imported from the Cote d'Ivoire, it appears more likely that this case reflects a relapse attributable to a failure to administer a radical cure, rather than to chloroquine resistance(10). In cases of indigenous vivax malaria, recurrence rates of approximately 3% after a standard course of treatment have been reported, but these failures appear to be the result of primaquine tolerance rather than chloroquine resistance. Within this context, our report describes the first case of chloroquine-resistant P. vivax infection in Korea.
Treatment failure of vivax malaria after chloroquine treatment is not necessarily indicative of chloroquine-resistant P. vivax infection; they can be caused by several factors, including, non-compliance or poor absorption of the medication, chloroquine resistance (i.e., recrudescence), primaquine resistance or tolerance, and reinfection(11). In our case, in which the administration of medication is strictly supervised, the possible causes are either inadequate absorption or chloroquine resistance. Measurement of blood level of chloroquine clearly separates the two conditions, however unfortunately, at the time in Korea, measurement of blood level of chloroquine is not a routine test. Because our case showed no obvious evidence of malabsorption or emesis, inadequate absorption of chloroquine could be excluded. A review literature shows that even 0.3 g of chloroquine can effectively cure blood-stage P. vivax(3). So the therapeutic dose of hydroxychloroquine over 48 hours in our patient could easily achieve minimally effective blood level of chloroquine, even if substantial degree of malabsorption might be present. In addition, satisfactory response with oral quinidine is another evidence of normal absorption of the medications. On the assumption that chloroquine is normally absorbed, our case meets the clinical criteria of chloroquine resistance, i.e., persistence of parasitemia for 4 days or recurrence of vivax malaria by within 35 days after the administration of chloroquine(3). Persistence of parasitemia for 4 days after administration of a therapeutic dose of chloroquine represents the highest grade resistance.
Several patterns of nucleotide polymorphism are associated with chloroquine-resistant malaria. In P. falciparum, two relevant proteins, Pgh1 and PfCRT, encoded by P. falciparum multidrug resistance gene 1 and P. falciparum chloroquine resistance transporter (pfcrt) respectively, have been identified as molecules that participate in chloroquine resistance(12, 13). As to P. vivax, resistance of P. vivax to antifolate agents is strongly associated with dhfr gene(6). Although the homologous genes of pfcrt and pfmdr1, pvcg10 and pvmdr1, were identified in P. vivax(14, 15), the molecular mechanism of chloroquine resistance in P. vivax is not clear and remains controversial. One report shows nucleotide polymorphisms at 2 codons of the pvmdr1 gene, i.e., Y976F and F1076L, in isolates from various areas including Indonesia(15), whereas the other report describes that there is no association with chloroquine resistance and mutation in pvmdr1 gene(7). Drawbacks of the above two studies are that the former identifies nucleotide polymorphism in isolates of which resistance to chloroquine is not confirmed, and the latter study uses ill-defined resistant isolates. In this context, our study shows that the results of gene sequencing revealed only one substitution located at position 1,076 (F→L) in the isolate definitely resistant to chloroquine, and suggests that the insignificant relationship of the pvmdr1 gene polymorphisms reported in the previous report(15) with chloroquine resistance in vivax malaria. The identified nucleotide polymorphism, F1076L, may be an incidental finding or a surrogate marker of resistance to other antimalarial agents, such as primaquine, or just a marker of molecular evolution and population diversity. Further cases will clarify our suggestion.
The optimal treatment of chloroquine-resistant vivax malaria remains debatable. Several therapies have proven effective, including, halofantrine, chloroquine plus primaquine, chloroquine plus doxycycline, Malarone® (250 mg of atovaquone and 100 mg of proguanil) plus primaquine, and sulfadoxine/pyrimethamine (42% response rate). Malarone alone has also proven effective as a prophylactic agent. In addition, several case reports support the notion that mefloquine or quinine followed by Fansidar also yield successful outcomes, but other reports have cited the failures of mefloquine and Malarone as prophylactic agents. Although its efficacy in cases of vivax malaria has not yet been thoroughly evaluated, our patient was treated with oral quinidine sulfate, as quinidine is believed to exhibit properties similar to those of quinine with regard to both its efficacy and its adverse effects(16). Moreover, oral quinidine is available in almost every Korean hospitals as an agent for use in the treatment of cardiac arrhythmia, and therefore can be accessed without delay in severe cases of malaria. Doxycycline is also known to be an effective treatment for vivax malaria, although the time required for the clearance of parasitemia by doxycycline treatment is approximately 150 hours. Further clinical studies are required in order to confirm the therapeutic efficacy of quinidine for the treatment of chloroquine- resistant P. vivax malaria.
Considering the increase in number of international travelers, especially to malaria endemic regions, imported malaria is an anticipated health problem of increasing importance. In the treatment of patients with imported malaria, the possibility of chloroquine resistance even in vivax malaria should be considered.
Figures and Tables
Figure 2
Deduced amino acid sequence of P. vivax pvmdr1 gene of the isolate from the case. The F1076L substitution was noticed.

References
1. Rieckmann KH, Davis DR, Hutton DC. Plasmodium vivax resistance to chloroquine. Lancet. 1989. 2:1183–1184.

2. Maguire JD, Sumawinata IW, Masbar S, Laksana B, Prodjodipuro P, Susanti I, Sismadi P, Mahmud N, Bangs MJ, Baird JK. Chloroquine-resistant Plasmodium malariae in south Sumatra, Indonesia. Lancet. 2002. 360:58–60.

3. Baird JK. Chloroquine resistance in Plasmodium vivax. Antimicrob Agents Chemother. 2004. 48:4075–4083.
4. Patsoula E, Spanakos G, Sofianatou D, Parara M, Vakalis NC. A single-step, PCR-based method for the detection and differentiation of Plasmodium vivax and P. falciparum. Ann Trop Med Parasitol. 2003. 97:15–21.

5. Figtree M, Pasay CJ, Slade R, Cheng Q, Cloonan N, Walker J, Saul A. Plasmodium vivax synonymous substitution frequencies, evolution and population structure deduced from diversity in AMA1 and MSP1 genes. Mol Biochem Parasitol. 2000. 108:53–66.

6. Na BK, Lee HW, Moon SU, In TS, Lin K, Maung M, Chung GT, Lee JK, Kim TS, Kong Y. Genetic variations of the dihydrofolate reductase gene of Plasmodium vivax in Mandalay Division, Myanmar. Parasitol Res. 2005. 96:321–325.

7. Sá JM, Nomura T, Neves J, Baird JK, Wellems TE, del Portillo HA. Plasmodium viavx: allele variants of the mdr1 gene do not associate with chloroquine resistance among isolates from Brazil, Papua, and monkey-adapted strains. Exp Parasitol. 2005. 109:256–259.

8. Yi KJ, Chung MH, Kim HS, Kim CS, Pai SH. A relapsed case of imported tertian malaria after a standard course of hydroxychloroquine and primaquine therapy. Korean J Parasitol. 1998. 36:143–146.

9. Na DJ, Han JD, Cha DY, Song IK, Choi HW, Chung EA, Park CW, Choi JS. Imported tertian malaria resistant to primaquine. Korean J Intern Med. 1999. 14:86–89.

10. Han SH, Byun DW, Chu WS, Woo JH, Hong ST. A case of recurrent Malaria-imported infection. Korean J Infect Dis. 1991. 23:125–129.
11. Collins WE, Jeffery GM. Primaquine resistance in Plasmodium vivax. Am J Trop Med Hyg. 1996. 55:243–249.

12. Foote SJ, Kyle DE, Martin RK, Oduola AM, Forsyth K, Kemp DJ, Cowman AF. Several alleles of the multidrug-resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum. Nature. 1990. 345:255–258.

13. Wellems TE, Walker-Jonah A, Panton LJ. Genetic mapping of the chloroquine-resistance locus on Plasmodium falciparum chromosome 7. Proc Natl Acad Sci USA. 1991. 88:3382–3386.

14. Nomura T, Carlton JM, Baird JK, del Portillo HA, Fryauff DJ, Rathore D, Fidock DA, Su X, Collins WE, McCutchan TF, Wootton JC, Wellems TE. Evidence for different mechanisms of chloroquine resistance in 2 Plasmodium species that cause human malaria. J Infect Dis. 2001. 183:1653–1661.

15. Brega S, Meslin B, de Monbrison F, Severini C, Gradoni L, Udomsangpetch R, Sutanto I, Peyron F, Picot S. Identification of the Plasmodium vivax mdr- like gene (pvmdr1) and analysis of single-nucleotide polymorphisms among isolates from different areas of endemicity. J Infect Dis. 2005. 191:272–277.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download