Abstract
Background
Polymerase-chain reaction (PCR) detection is useful to diagnosis of pertussis at initial stage because the growth rate of Bordetella pertussis (B. pertussis) is relatively slow. Currently, the primer set for the insertion sequence IS481 (BP primer) is used widely for PCR detection of B. pertussis. However, the cross-reactivity of BP primer set with Bordetella holmesii (B. holmesii) was reported recently. Therefore, discrimination of B. pertussis and B. holmesii is needed in PCR step. For this reason, we developed new primer sets based on 16S rDNA sequence for diagnostic use and estimated the efficiency of these new primer sets.
Materials and Methods
The specific PCR primers were designed from the aligned sequence matrix of 16S rDNA genes of various Bordetella species. The specificity of designed primers were estimated using clinically important 4 Bordetella species, B. pertussis, B. holmesii, Bordetella parapertussis (B. parapertussis) and Bordetella bronchiseptica (B. bronchiseptica). The sensitivity to B. pertussis of designed primers was also estimated and compared with BP primer set.
Results
As the results, the developed new primer set successfully distinguished B. pertussis and other Bordetella species containing B. holmesii. In the sensitivity assay, the detectable limits of 16S-F2/16S-R1 primer set for B. pertussis were revealed as 5 pg of genomic DNA and 105 cells/mL of cell suspension. In addition to these, identical results between BP with primer and new primer were obtained in clinical samples.
Pertussis is one of the respiratory diseases caused by bacterial infection and specifically severe in the infants. The high global incidences of pertussis were reported until 1980's (1,982,384 cases/year)(1) but they were greatly reduced by vaccination campaign, which started from 1940. However, local outbreaks of pertussis were reported between 1990-2000 in well-vaccinated countries like USA, Netherlands, Japan, and Australia(2-4). Although there were many efforts to find out the reason of these outbreaks, they were not yet found. Therefore, to prevent the disease from spreading in early stage, the diagnostic process plays an important role.
Usually, the diagnostic gold standard is to confirm the pathogen by culture. However, the growth rate of Bordetella pertussis, the pathogen of pertussis, is quite slow. For this reason, the PCR detection is a critical step to determine the result in early phase. Until now, various kinds of diagnostic primer sets were developed. From these primer sets, BP1/2 for IS481 element(5, 6), PTp1/ p2 for pertussis toxin promoter region(5, 7, 8) and P1/2/3 for porin gene upstream region(9) are common primer sets used in clinical application. Especially, because of its high sensitivity and specificity, BP primer set is most widely used primer. However, despite of high sensitivity of BP primer, the cross-reactivity of BP primer set to B. holmesii was recently reported(10-13).
B. holmesii is one of the species of Bordetella Genus and shows highest similarity to B. pertussis in 16S rDNA sequence analysis(14). There were few reports about the pathogenicity and infectivity of B. holmesii for human, this strain was isolated from the nasopharyngeal specimens of patients with pertussis-like symptoms(14, 15). Moreover, the specific primer for B. holmesii was not reported yet and BP primers are currently applied to B. holmesii detection(10, 12, 13). Therefore, in diagnostic PCR diagnosis step, discriminative analysis between B. pertussis and B. holmesii is required.
For this reason, we designed new diagnostic primer sets for 4 major species of Bordetella Genus based on 16S rDNA sequence and the efficiency of designed primer sets was estimated.
The reference bacterial strains used in this study were Bordetella pertussis (ATCC 9797), Bordetella holmesii (ATCC 51541), Bordetella parapertussis (ATCC 15237) and Bordetella bronchiseptica (ATCC 31124). In addition to these strains, the nasopharyngeal specimens collected from pertussis patients were also used as confirmation test.
The genomic DNAs of 4 Bordetella species were purified from cultured cells and used as PCR templates. For this, all of strains were incubated on Regan-Lowe medium without cephalexin for 5-7 days at 37°. After incubation, the bacterial cells were collected by scrapping and genomic DNA was isolated using commercial kit according to the manufacturer's instruction (QIAamp DNA Mini Kit, QIAGEN).
For clinical specimens, collected nasopharyngeal aspirates (NPA) were directly used as templates for PCR reaction. Some portion (0.5-1 mL) of the specimens was heated for 5 min in boiling water. Then after centrifugation, 1-2 µL of supernatant was used as PCR template.
The 4 PCR primers were designed from 16S rDNA sequences of 4 Bordetella species (B. holmesii, B. bronchiseptica, B. pertussis and B. parapertussis). The DNA sequence information of 16S rDNA gene was retrieved from NCBI GenBank. These collected nucleotide sequences were aligned using multiple alignment method by MEGA program(16) and the sequence similarity of 16S rDNA gene was calculated by p-distance method at Family level and Genus level(17). From these aligned data matrix, the region containing different nucleotide was analyzed by Sequence Output program and the PCR primers were designed by these differences (Fig. 1). For reference, the BP primers sequence (BP1:5'-GAT TCA ATA GGT TGT ATG CAT GGT T-3', BP2:5'-TTC AGG CAC ACA AAC TTG ATG GGC G-3') was cited from the published paper(12).
The standard reaction mixture (20 µL) was composed of 2 µL of 10x buffer, 0.5 µL of dNTPs mix (2.5 mM each), 1 µL of F-primer (5 pmoL/µL), 1 µL of R-primer (5 pmoL), 0.2 µL of SP-Taq polymerase (2.5 Unit/µL), 1 µL of template (purified genomic DNA or bacterial suspension) and distilled water was added to make total 20 µL of reaction volume. For 16S primer sets, dimethylsulfoxide (DMSO) was added to make 2.5% of final concentration.
The temperature condition for new 16S primer sets was 35 cycles of denaturation at 98° for 10 sec and annealing/extension at 64° or 66° for 15 sec according to primer sets. In the case of BP primer sets, 35 cycles of denaturation at 95° for 5 sec and annealing/extension at 54° for 10 sec were performed.
The cross reactivity of BP primer to B. holmesii and other 2 Bordetella species (B. bronchiseptica and B. parapertussis) was confirmed in this study. As shown in Fig. 2, positive bands were appeared in both lanes (lane 4 and 5) of B. pertussis and B. holmesii. However, B. parapertussis (lane 2) and B. bronchiseptica (lane 3) were not detected by BP primers set. Therefore, the use of BP primers set in PCR diagnosis of pertussis should be considered.
In this study, we considered the 16S rDNA gene to develop new primer set for PCR-diagnosis of pertussis. For this, the 16S rDNA sequences of 4 major Bordetella species (B. pertussis, B. holmesii, B. bronchiseptica and B. parapertussis) were collected from NCBI GenBank and aligned by multiple alignment method to find sequence variations. As the result, there were no specific variable regions. This indicates that 16S rDNA sequence was highly conserved in Bordetella species (98.7% of sequence similarity). However, the two positions of 661 bp and 782 bp in alignment matrix showed specificity according to their species. We selected these two sites as the motif for PCR primer design (Fig. 1) and 4 new primers were designed. As shown in Fig. 1, the designed primers were different in only one nucleotide of their 3'-termini.
As mentioned above, the specificity of designed new PCR primer sets was examined to 4 Bordetella species. For specific amplification, the combinations of these 4 primers were applied. 16S-F1/16S-R1 set for B. holmesii, 16S-F2/16S-R1 set for B. pertussis, and 16S-F2/ 16S-R2 set for other Bordetella species (Fig. 1) were used. As the result, the specific amplification of B. pertussis, B. parapertussis and B. bronchiseptica using the combinatorial primers sets was successfully performed. As shown in Fig. 3, the positive PCR bands appeared in only each target species (Lane 2, Lane 5, Lane 11 and Lane 12). Therefore, it was confirmed that the designed PCR primer sets showed the specificity to each Bordetella species.
16S-F2 and 16S-R1 primers were applied for B. pertussis. As shown in Fig. 3, the positive band was detected only in B. pertussis sample (Lane 5). However, we confirmed that the non-specific band was also detectable in the sample of B. holmesii under mild PCR condition. Therefore, stringent PCR condition was required in the use of 16S-F2 and 16S-R1 primers. For this reason, we added dimethylsulfoxide (DMSO) to reaction mixture until 2.5% of final concentration. As the result, 16S-F2/16S-R1 primers set specifically detected B. pertussis in this stringent condition.
The sensitivity of these 2 primer sets (BP and 16S) for B. pertussis was estimated and compared. Under fixed PCR condition, the detection limit of target band was examined using purified genomic DNA. As shown in Fig. 4, when genomic DNA was used, the detection limit was 5 pg/reaction ((A)-lane 2). This estimated value was lower than the result of BP primer set (Fig. 4-(B)). BP gene was detectable as low as 10 fg of genomic DNA ((B)-lane 6).
In the case of cell suspension, as shown in Fig. 5, the detection limit of 16S rDNA gene was > 105 cells/ mL ((A)-lane 5). This result is not quite different with the result of BP gene. The detectable PCR band for BP gene was also shown from the cell suspension of > 104 cells/ml ((B)-lane 7). Therefore, these results indicated that 16S rDNA gene and BP gene showed similar detection range under practical diagnostic condition.
Applicability of new 16S primers to clinical specimens were estimated. The specimens were the nasopharyngeal aspirates collected from the suspected patients pertussis in 2006. These specimens were already confirmed positive for B. pertussis by BP primer PCR and culture. As shown in Fig. 6, all positive specimens were also confirmed positive by 16S PCR primer (Lane 1-4 in Fig. 6). Therefore, the newly developed 16S PCR primer sets were also applicable to the clinical specimens.
According to the WHO guide for Bordetella species diagnosis(1), the cross reactivity of BP primer to B. holmesii was already described. However, BP primer set is still used in most diagnostic laboratories because of its high sensitivity and specificity. Moreover, uncertainty of B. holmesii for human infection and carriage was also the reason. However, pathogenicity and carriage activity of B. holmesii for human were also proved recently(14, 15). For these reasons, there is need to develop new primer sets for discrimination of these two Bordetella species at PCR diagnostic step.
For this, we considered the 16S rDNA gene. As already known, 16S rDNA gene is most frequently used taxonomic marker in all Bacteria and Archaea(18). Firstly, we confirmed the sequence similarity of 16S rDNA gene to estimate their usefulness for B. pertussis. The group mean similarity of Bordetella Genus was 98.7% and it was higher than overall mean similarity of Alcaligenaceae Family (93.6%) containing Bordetella Genus. This indicates that 16S rDNA sequence is highly conserved to each Genus of Alcaligenaceae Family and this gene is a useful taxonomic marker to discriminate each Genus of Alcaligenaceae Family.
Currently, 9 Bordetella species have been reported in NCBI Taxonomy database (B. ansorpii, B. avium, B. bronchiseptica, B. hinzii, B. holmesii, B. parapertussis, B. petrii, B. trematum, B. pertussis). Among these species, the detectable species from human respiratory specimens were B. bronchiseptica, B. hinzii, B. holmesii, B. parapertussis and B. pertussis(19-21). Because B. hinzii was reported as rarely isolated species from immunocompromised(20) and cystic fibrosis patients(22), we did not considered it as major Bordetella species isolated from normal respiratory specimens. However, discrimination of B. pertussis and B. hinzii would be also possible by using new 16S primer sets (Fig. 1).
Although B. ansorpii and B. trematum were also isolated from human originated specimens(23, 24), they did not relate with human respiratory infection. Therefore, 4 Bordetella species (B. ansorpii, B. petrii, B. trematum, B. avium) were excluded from diagnostic targets in this study.
Especially, we also confirmed the specific primer set (16S-F1 and 16S-R1) for B. holmesii in this study (Fig. 3, Lane (2)). Until now, biochemical tests and cellular fatty acids analysis were mainly performed as identification tests for B. holmesii(14). In the molecular methods, DNA hybridization technique and 16S rDNA sequence analysis were performed(21, 25). However, there is no method for rapid identification of B. holmesii. IS481 based BP primer set for B. pertussis is used in B. holmesii detection(10, 12, 13). Therefore, there is need to develop PCR primer of B. holmesii for rapid identification. Although recA gene was recently reported as target gene for specific PCR of B. holmesii(26), other specific primer or target gene were not reported. As the result, the confirmed PCR primer set for B. holmesii in this report will be applicable to clinical diagnostic tests.
As shown in Fig. 4, 16S primer set showed 100 times lower sensitivity than BP primer set. It is attributed in the copy numbers of these two related genes. The BP primer was originated in IS481 repetitive sequence. It was known that there were approximately 80-100 copies of IS481 region in a bacterial cell(12, 27). However, for the 16S rDNA gene in the case of Tohama I strain, 3 copies per genome were present(28). Therefore, the major reason of reduced sensitivity of 16S primer was inferred as this different copy number.
As the conclusion, the specific PCR primers sets for B. pertussis, B. holmesii, B. parapertussis and B. bronchiseptica were developed based on 16S rDNA gene and their specific amplification of target strain was successfully confirmed in this study. Although the sensitivity was relatively low as compared to BP primers set, these new primer sets are applicable to clinical specimens and especially useful to discriminate B. holmesii and B. pertussis.
Figures and Tables
Fig. 1
Design of PCR primers. (A) Multiple alignment of 16S rDNA sequences from B. holmesii, B. bronchiseptica, B. pertussis, B. parapertussis and B. hinzii. (B) Designed PCR primers sequences and specificity by combination of primers. The 16S rDNA sequences of indicated Bordetella strains were retrieved from NCBI GenBank and the collected sequences were aligned by multiple alignment method. From the generated data matrix (A), the variable sites were searched and 4 PCR primers were designed. The specificity for each target species was derived from the combinational sets of 4 primers, as shown in (B).

Fig.2
The cross reactivity of BP primer set between B. holmesii and B. pertussis. Lane M, Molecular size marker (100 bp ladder); Lane 1, Negative control; Lane 2, B. parapertussis; Lane 3, B. bronchiseptica; Lane 4, B. pertussis; Lane 5, B. holmesii. The genomic DNAs of 4 Bordetella species were purified with QIAamp DNA Mini Kit and 10 ng of purified genomic DNAs were added to PCR reaction mixture. The temperature condition was 35 cycles of denaturation at 95° for 5 sec and annealing/extension at 54° for 10 sec. After reaction, the amplified target band was confirmed by 1.5% agarose gel electrophoresis.
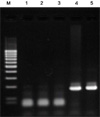
Fig.3
The specific amplification of 16S rDNA using newly designed 16S primer sets. Lane M, Molecular size marker (100 bp ladder); Lane 1, 5, 9, B. pertussis; Lane 2, 6, 10, B. holmesii; Lane 3, 7, 11, B. parapertussis; Lane 4, 8, 12, B. bronchiseptica. The combined primer sets (Fig. 1) were tested using purified genomic DNA of 4 Bordetella species (B. pertussis, B. holmesii, B. parapertussis and B. bronchiseptica). 5 ng of purified genomic DNAs were used in PCR reaction and 2.5% dimethylsulfoxide (final concentration) was added to reaction mixture. The temperature condition also differently applied to primer sets. For 16S-F1/R1, 35 cycles of denaturation at 98° for 10 sec and annealing/extension at 64° for 15 sec were performed. For 16S-F2/R1 and 16S-F2/R2, the temperature condition was 35 cycles of denaturation at 98° for 10 sec and annealing/ extension at 66° for 15 sec. After reaction, the amplified target band was confirmed by 1.5% agarose gel electrophoresis.

Fig.4
Comparison of the sensitivity for detection of B. pertussis using purified genomic DNA by 16S and BP primer sets. (A) Sensitivity of 16S primer for detection of B. pertussis. Lane M, Molecular size marker (100 bp ladder); Lane 1, : 1 pg; Lane 2, 5 pg Lane 3, 10 pg; Lane 4, 50 pg; Lane 5, 100 pg; Lane 6, 500 pg; Lane 7, 1 ng. (B) Sensitivity of BP primer for detection of B. pertussis. Lane M, Molecular size marker (100 bp ladder); Lane 1, 1 ng; Lane 2, 100 pg; Lane 3, 10 pg; Lane 4, 1 pg; Lane 5, 0.1 pg; Lane 6, 0.01 pg; Lane 7, 0.001 pg; Lane 8, Negative control. The sensitivity of 16S and BP primer sets were estimated using various concentration of purified B. pertussis genomic DNA. As indicated in this fig, 0.001 pg - 1 ng of purified genomic DNA was tested. After reaction, 10 µL of reactants were electrophoresed on 1.5% agarose gel and the amplified band was observed under UV illuminator.

Fig.5
Comparison of the sensitivity for detection of B. pertussis using cell suspension by 16S and BP primer sets. (A) Sensitivity of 16S primer for detection of B. pertussis. Lane M, Molecular size marker (100 bp ladder); Lane 1, 1010 cells/mL; Lane 2, 109 cells/mL; Lane 3, 108 cells/mL; Lane 4, 107 cells/mL; Lane 5, 106 cells/mL; Lane 6, 105 cells/mL; Lane 7, 104 cells/mL. (B) Sensitivity of BP primer for detection of B. pertussis. Lane M, Molecular size marker (100 bp ladder); Lane 1, Negative control; Lane 2, 1010 cells/mL; Lane 3, 109 cells/mL; Lane 4, 108 cells/mL; Lane 5, 107 cells/mL; Lane 6, 106 cells/mL; Lane 7, 105 cells/mL; Lane 8, 104 cells/mL. sensitivity of 16S and BP primer sets were estimated using various concentrations of cell suspensions. As indicated in this fig, 104-1010 cells/mL of cell suspensions were heated in boiled water for 5 min and centrifuged at 15,000 rpm for 5 min. After centrifugation, 1 µL of supernatant was added to PCR reaction mixture as template. The amplified PCR band was observed under UV illuminator after electrophoresis on 1.5% agarose gel.

Fig.6
Application of 16S PCR primer (16S-F2/R1) to clinical specimens. Lane M, Molecular size marker (100 bp ladder); Lane 1-4, Pertussis positive specimens; Lane 5-6, Pertussis negative specimens. The clinical specimens were nasopharyngeal aspirates collected from pertussis-like patients. 0.5 mL of the collected specimens was heated in boiled water for 5 min and centrifuged at 15,000 rpm for 5 min. After centrifugation, 1 µL of supernatant was added to PCR reaction mixture as template. The amplified PCR band was observed under UV illuminator after electrophoresis on 1.5% agarose gel. All positive and negative specimens were already confirmed by BP primer set.
References
1. WHO. WHO/IVB/04.14. Laboratory manual for the diagnosis of whooping cough caused by Bordetella pertussis/Bordetella parapertussis. 2004.
2. de Melker HE, Conyn-van Spaendonck MA, Rumke HC, van Wijngaarden JK, Mooi FR, Schellekens JF. Pertussis in the netherlands: an outbreak despite high levels of immunization with whole-cell vaccine. Emerg Infect Dis. 1997. 3:175–178.

3. de Melker HE, Schellekens JFP, Neppelenbroek SE, Mooi FR, Rumke HC, Conyn-van Spaendonck MAE. Reemergence of pertussis in the highly vaccinated population of the Netherlands: Observations on surveillance data. Emerg Infect Dis. 2000. 6:348–357.

4. Mooi FR, van Loo IH, King AJ. Adaptation of Bordetella pertussis to vaccination: A cause for its reemergence? Emerg Infect Dis. 2001. 7:suppl 3. 526–528.

5. Fry NK, Tzivra O, Li YT, McNiff A, Doshi N, Maple PA, Crowcroft NS, Miller E, George RC, Harrison TG. Laboratory diagnosis of pertussis infections: the role of PCR and serology. J Med Microbiol. 2004. 53:519–525.

6. van der Zee A, Agterberg C, Peeters M, Schellekens J, Mooi FR. Polymerase chain reaction assay for pertussis: simultaneous detection and discrimination of Bordetella pertussis and Bordetella parapertussis. J Clin Microbiol. 1993. 31:2134–2140.

7. Birkebaek NH, Heron I, Skjodt K. Bordetella pertussis diagnosed by polymerase chain reaction. APMIS. 1994. 102:291–294.
8. Houard S, Hackel C, Herzog A, Bollen A. Specific identification of Bordetella pertussis by the polymerase chain reaction. Res Microbiol. 1989. 140:477–487.

9. Li Z, Jansen DL, Finn TM, Halperin SA, Kasina A, OConnor SP, Aoyama T, Manclark CR, Brennan MJ. Identification of Bordetella pertussis infection by shared-primer PCR. J Clin Microbiol. 1994. 32:783–789.

10. Loeffelholz MJ, Thompson CJ, Long KS, Gilchrist MJ. Detection of Bordetella holmesii using Bordetella pertussis IS481 PCR assay. J Clin Microbiol. 2000. 38:467.

11. Poddar SK. Detection and discrimination of B pertussis and B. holmesii by real-time PCR targeting IS481 using a beacon probe and probe-target melting analysis. Mol Cell Probes. 2003. 17:91–98.

12. Reischl U, Lehn N, Sanden GN, Loeffelholz MJ. Real-Time PCR assay targeting IS481 of Bordetella pertussis and molecular basis for detecting Bordetella holmesii. J Clin Microbiol. 2001. 39:1963–1966.

13. Templeton KE, Scheltinga SA, van der Zee A, Diederen BM, van Kruijssen AM, Goossens H, Kuijper E, Claas EC. Evaluation of real-time PCR for detection of and discrimination between Bordetella pertussis, Bordetella parapertussis, and Bordetella holmesii for clinical diagnosis. J Clin Microbiol. 2003. 41:4121–4126.

14. Yih WK, Silva EA, Ida J, Harrington N, Lett SM, George H. Bordetella holmesii-like organisms isolated from Massachusetts patients with pertussis-like symptoms. Emerg Infect Dis. 1999. 5:441–443.

15. Mazengia E, Silva EA, Peppe JA, Timperi R, George H. Recovery of Bordetella holmesii from patients with pertussis-like symptoms: use of pulsed-field gel electrophoresis to characterize circulating strains. J Clin Microbiol. 2000. 38:2330–2333.

16. Kumar S, Tamura K, Nei M. MEGA3: Integrated Software for Molecular Evolutionary Genetics Analysis and Sequence Alignment. Brief Bioinform. 2004. 5:150–163.

17. Nei M, Kumar S. Molecular Evolution and Phylogenetics. 2000. New York: Oxford University Press.
18. Mattoo S, Cherry JD. Molecular pathogenesis, Epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin Microb Rev. 2005. 18:326–382.

19. Cummings CA, Brinig MM, Lepp PW, van de Pass , Relman DA. Bordetella species are distinguished by patterns of substantial gene loss and host adaptation. J Bacteriol. 2004. 186:1484–1492.

20. Vandamme P, Hommez J, Vancanneyt M, Monsieurs M, Hoste B, Cookson B, Wirsing von Konig CH, Kerster K, Blackall PJ. Bordetella hinzii sp. Nov., isolated from poultry and humans. Int J Syst Bacteriol. 1995. 45:37–45.

21. Weyant RS, Hollis DG, Weaver RE, Amin MF, Steigerwalt AG, O'Connor SP, Whitney AM, Daneshvar MI, Moss CW, Brenner DJ. Bordetella holmesii sp. Nov., a new gram-negative species associated with septicemia. J Clin Microbiol. 1995. 33:1–7.

22. Coenye T, Goris J, Spilker T, Vandamme P, LiPuma JJ. Characterization of unusual bacteria isolated from respiratory secretions of cystic fibrosis patients and description of Inquilinus limosus gen. Nov., sp. Nov. J Clin Microbiol. 2002. 40:2062–2069.

23. Ko KS, Peck KR, Oh WS, Lee NY, Lee JH, Song JH. New species of Bordetella, Bordetella ansorpii sp. Nov., isolated from the purulent exudate of an epidermal cyst. J Clin Microbiol. 2005. 43:2516–2519.

24. Vandamme P, Heyndrickx M, Vancanneyt M, Hoste B, De Vos P, Falsen E, Kersters K, Hinz KH. Bordetella trematum sp. nov., isolated from wounds and ear infections in humans, and reassessment of Alcaligenes denitrificans Ruger and Tan 1983. Int J Syst Bacteriol. 1996. 46:849–858.

25. Shepard CW, Daneshvar MI, Kaiser RM, Ashford DA, Lonsway D, Patel JB, Morey RE, Jordan JG, Weyant RS, Fischer M. Bordetella holmesii Bacteremia: A newly recognized clinical entity among asplenic patients. Clin Infect Dis. 2004. 38:799–804.

26. Antila M, He Q, de Jong C, Aarts I, Verbakel H, Bruisten S, Keller S, Haanpera M, Makinen J, Eerola E, Viljanen MK, Mertsola J, van der Zee A. Bordetella holmesii DNA is not detected in nasopharyngeal swabs from Finnish and Dutch patients with suspected pertussis. J Med Microbiol. 2006. 55:1043–1051.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download