Abstract
The present study was done to scrutinize the possible relation between infective genes and antimicrobial resistance in Enterococcus faecalis and Enterococcus faecium. Considering the fact that the presence of recognized infective determinants among clinical isolates may promote the emergence of infections and persistence of Enterococci in hospital settings, which can lead to an increase in antimicrobial resistance. 175 E. faecalis and 67 E. faecium isolated from clinical specimens were used. The isolates were identified, and then antibiotic susceptibility testing was performed. The MIC of vancomycin and teicoplanin were determined by broth microdilution method. The presence of infective genes esp, hyl and asa1 was scrutinized using PCR. Of the 280 enterococcal isolates, 175 (62.5%) isolates were identified as E. faecalis, 67 (24%) as E. faecium and 38 (13.5%) as Enterococcus spp. The results of the antibiotic susceptibility testing showed resistance rates of 5% and 73% to vancomycin and teicoplanin in E. faecalis and E. faecium isolates, respectively. The statistical analysis showed that the esp infective gene has significant associations with ciprofloxacin, erythromycin and tetracycline in E. faecium and with chloramphenicol in E. faecalis strains; the hyl with teicoplanin and vancomycin in E. faecium strains; and also asa1 with vancomycin in E. faecium and with ampicillin and chloramphenicol in E. faecalis strains. Regarding the relationships between virulence genes and antibiotic resistance in strains of E. faecalis and E. faecium, detection of infective factors associated with invasive diseases has become a major issue of concern.
Enterococci bacteria are facultative anaerobic grampositive cocci, which are considered part of the normal flora in humans and animals. However, these microorganisms may be the cause of several serious systematic infections too.123
The two most common Enterococcus species, E. faecalis and E. faecium are responsible for 80-90% and 5-10% of human enterococcal infections, respectively.2 E. faecalis is the most common isolate of nosocomial infections, but newly, due to increasing resistance to some antimicrobial agents, especially vancomycin, E. faecium isolates are also being considered.4 It has been shown that separate lineages of E. faecalis and E. faecium are leading causes of the large number of the multidrug-resistant enterococcal infections. According to several investigations, CC17 and closely related strains are the main agents of most hospital-acquired infections association with E. faecium.5 Although these organisms lack strong virulence factors, they may have an innate resistance/tolerance to many important antibacterial agents such as, cephalosporins, cotrimoxazole, and low levels of penicillin and aminoglycosides, polymyxin, lincosamide, trimethroprim-sulfamethoxazole, monobactams, streptogramin. They are also able to acquire resistance to penicillin, chloramphenicol, tetracyclines, aminoglycosides and vancomycin.3678
Pathogenicity and increased risk of acquisition of enterococcal infections is linked to antimicrobial resistance and expression of virulence factors including, adhesion factors, translocation, and immune evasion. In hospital settings, infective potential of enterococci can be due to selective the advantages conferred by their antibiotic resistance. Furthermore, there is great concern about infectious diseases because of spreading antimicrobial resistance genes. The most important infective agents of enterococci which have been identified, include: aggregation substances (asal), cytolysin (cyl), hyaluronidase (hyl), the enterococcal surface protein (esp) and gelatinase (gelE).910 It has been found that some of virulence factors such as, agg, esp and cyl genes, located on 153-kb pathogenicity island.11 Persistence of enterococci in the hospital setting may be associated with their virulence factors.11 Based on several studies, the virulence factors of gelatinase, aggregation substance and cytolysin have not been found in E. faecium in contrast to E. faecalis. On the other hand, esp and hyl have been found in both E. faecalis and E. faecium.12
Furthermore, clinical isolates of enterococci have virulence determinants that may result in promoting the emergence of infections and the persistence of these organisms in hospital settings which consequently can lead to increased resistance.13 This study was designed to scrutinize some virulence genes including asa1 (aggregation substance), esp (enterococcal surface protein), gelE (gelatinase), hyl (hyaluronidase) in clinical isolates of E. faecalis and E. faecium and to investigate possible correlations between virulence and antibiotic resistance.
One hundred and seventy-five E. faecalis and sixty-seven E. faecium strains were collected from discrete clinical samples submitted to three teaching hospitals (including Beheshti, Besat and Farshchian Hospitals) located in Hamedan, Iran, from December 2012 to May 2014. The origins of the isolates were as follows: urine 200 (82.6%), endotracheal aspirate 23 (9.5%), blood 8 (3.3%), Skin soft tissue 6 (2.5%), and body fluids 5 (2.1%). The isolates were pinpointed using routine microbiological methods.14 Then, PCR targeting D-alanine- D-alanine ligases for E. faecalis (ddl E. faecalis) and E. faecium (ddl E. faecium) was used to confirm phenotypic speciation.15
Firstly, enterococcal DNA was extracted by boiling.16 Then, a mastermix PCR Kit [(PCR 2X Taq premix Mastermix), Ariatous Biotec Co.] was used to perform the PCR reaction. PCRs were performed with specific primers for each gene (Table 1) with some modifications to Kariyama's protocol15 using Eppendorf and Biorad thermocycler in a final volume of 20 µL. The thermal cycle program was performed by initial denaturation at 95℃ for 5 min, followed by amplification in 30 cycles of denaturation at 95℃ for 30 s, annealing at 52.5℃ for 30 s and elongation at 72℃ for 1 min, and a final extension at 72℃ for 10 min. E. faecalis ATCC 29212 and E. faecium BM4147 were used as quality control strains.
The antimicrobial susceptibilities of 175 E. faecalis and 67 E. faecium strains were examined by using the disk agar diffusion (DAD) method in accordance with the Clinical and Laboratory Standards Institute (CLSI) guidelines.1718 Erythromycin (15 µg), Tetracycline (30 µg), Ciprofloxacin (5 µg), Vancomycin (30 µg), Teicoplanin (30 µg), Norfloxacin (10 µg), Nitrofurantoin (300 µg), Quinopristin-Dalfopristin [Synercid (15 µg)] (Mast Co., UK), Chloramphenicol (30 µg), Gentamicin (10 µg), Linezolid (30 µg), and Ampicillin (10 µg) (HiMedia Mumbai Co., India) were used for antimicrobial susceptibility testing (AST).
In addition, minimum inhibitory concentrations (MIC) of the glycopeptide antibiotics i.e. vancomycin and teicoplanin (Sigma-Aldrich, Poole, Co., UK) against the E. faecalis and E. faecium isolates were determined using the microdilution broth method.1718 E. faecalis ATCC 29212 (Vancomycin sensitive), E. faecalis ATCC 51299 (vanB positive), E. faecalis E206 (vanA positive) were used as quality control.
Multiplex PCR and single PCR were used for the identification of esp, asa1 and hyl virulence determinants using specific primers for each gene with some modifications on Vankerckhoven's protocol (Table 1).1319 Briefly, the final volume multiplex PCR reaction for genes esp and asa1, was 25 µL and for hyl gene was 20 µL. The PCR reactions were done for both mixtures on a Eppendorf and Biorad thermocycler (ASTEC Co., Japan) with an initial denaturation at 95℃ for 10 min, 30 cycles of amplification (denaturation at 94℃ for 1 min, annealing at 56℃ for 1 min, and extension at 72℃ for 1 min), and a final extension at 72℃ for 10 min.13 The E. faecalis ATCC 29212 (asa1 positive), E. faecium C68 (hyl and esp positive) were used as quality control.
Correlation between antibiotic susceptibility patterns and occurring virulence genes was analyzed statistically using the Chi-Square test; the differences were considered significant for p<0.0012 using the Bonferroni correction based on several primary comparisons. In addition, significant differences for simultaneous occurrance of virulence genes in enterococci strains was determined using Fisher's Exact test with a significance level of p<0.05. All tests were performed using SPSS software (version 19).
Using biochemical methods, of the 280 enterococcal isolates, 190 (67.8%), 75 (26.8%) and 15 (5.4%) isolates were recognized as E. faecalis, E. faecium and Enterococcus spp., respectively. Among the determined presumptive E. faecalis, E. faecium isolates, 175 (62.5%) E. faecalis and 67 (24%) E. faecium strains were confirmed using the PCR method (Fig. 1). Therefore, a total of 38 strains (13.5%) remained as a part of the Enterococcus genus and were excluded from the current study. Urine samples were the highest isolation source of E. faecalis and E. faecium strains. Among 175 E. faecalis strains, 153 isolates and among 67 E. faecium strains, 48 isolates were isolated from urine samples; followed by tracheal sample with 17 isolates (11 strains were determined as E. faecalis and 6 strains as E. faecium). The highest prevalence of E. faecium strains was observed in wounds and other organs samples (abscess and pulmonary secretions); among 12 strains isolated from wounds and other organs, 8 strains belonged to E. faecium (Table 2).
Table 3 shows the susceptibility patterns of 67 E. faecium and 175 E. faecalis strains to 12 commonly used antibiotics. Resistance to the majority of antibiotics except for chloramphenicol, tetracycline, and quinopristin-dalfopristin was higher in E. faecium isolates compared to E. faecalis isolates. However, they both showed good rates of sensitivity to linezolid (100%), nitrofurantoin and chloramphenicol (74.6%). In fact, no resistance to the linezolid antibiotic was observed among E. faecalis and E. faecium strains; and all isolates of E. faecalis were susceptible to nitrofurantoin. Among 175 E. faecalis strains, the greatest resistance was observed for Synercid antibiotic with frequency of 167 (95.4%) isolates; followed by tetracycline with 154 (88%), erythromycin with 109 (62.3%), ciprofloxacin with 69 (39.4%), gentamicin with 63 (36%), Norfloxacin with 58 (33%), chloramphenicol with 57 (32.6%), vancomycin and teicoplanin with 9 (5%) and ampicillin with 6 (3.4%) isolates. Among 67 E. faecium isolates, the greatest resistance was observed for gentamicin with a frequency of 62 (92.5%) strains, followed by erythromycin with 58 (86.6%), norfloxacin with 56 (83%), ciprofloxacin with 54 (80.6%), vancomycin with 51 (76%), tetracycline and teicoplanin with 49 (73%), Synercid with 47 (70%), ampicillin with 42 (62.7%), nitrofurantoin with 17 (25.4%), and chloramphenicol with 7 (10.4%).
Of 175 E. faecalis isolates, resistances of 9 (5%) and sensitivity of 166 (95%) isolates to vancomycin and teicoplanin were confirmed by the microdilution broth method. Of 67 E. faecium isolates, 51 strains (76%) were resistant to vancomycin using the disk diffusion method, but resistance of 49 strains (73%) to vancomycin was confirmed by the Microdilution Broth method. 2 strains (3%) of E. faecium that were determined as resistant strains by disk diffusion using the Microdilution Broth method were identified as intermediate strains. In addition, resistance of 49 (73%) and sensitivity of 18 (27%) E. faecium isolates to teicoplanin using disk diffusion method were confirmed by the microdilution broth method. All 9 vancomycin-resistant E. faecalis (VREfs) and 49 vancomycin-resistant Enterococcus faecium (VREfm) strains had high-level resistances to vancomycin and teicoplanin, concurrently. The MIC value among 9 VREfs strains and VREfm strains was ≤512, 512.
The distribution of infective factors among E. faecalis and E. faecium strains are presented in Table 4 and (Fig. 2 and 3). Among the E. faecalis strains, the asa1 gene was the most prevalent factor (97%), followed by the esp (78.3%) and hyl genes (56.6%); additionally, in E. faecium strains, the asa1 gene had the highest prevalence (100%) and hyl gene has the lowest frequency (71.6%), followed by the esp gene (82%).
In a total of 242 Enterococci strains, the asa1 gene was the most prevalent factor (98%), followed by the esp (79.3%) and hyl genes (60.7%). In the current study, the esp gene was found in 75% of the strains isolated from blood samples and 83% of strains isolated from wound infections.
Using statistical analysis, there was no significant difference for simultaneous occurrence of virulence genes in studied enterococci strains (p>0.05).
In the present study, we assessed antimicrobial resistance and three infective factors in clinically isolated E. faecalis and E. faecium and further analysis was conducted to scrutinize the relationship between the presence of virulence factors and antimicrobial resistances. In this study, there was a slight discrepancy between biochemical and PCR results for species identification, which is in accordance with other studies.2021 The main reason for this discrepancy is the similarities between Enterococcus species and the high phenotypic variation within individual species. PCR is a more accurate technique in comparison to the biochemical approach, so PCR results were preferred for the strains with discrepant identification.22 In this study, the esp gene was detected in 78.3% of E. faecalis and 82% of E. faecium isolates, this finding is similar to the results of other studies, which identified the esp gene in 47.1%,23 73%,24 68.4%7 of E. faecalis, and 80%,2526 65%,27 66%,23 71%,24 75%12 of E. faecium strains. However, this is in contrast to the findings of Shankar et al.28 and Channaiah et al.29 that reported the absence of esp in E. faecium. Although, as illustrated by Shankar et al.28 the esp gene was detected only in E. faecalis strains and other available studies have demonstrated a higher prevalence of the esp gene in E. faecalis,16 however, a study on the food and medical isolates indicated an increasing incidence of esp in clinical E. faecium isolates compared to E. faecalis.25
Willems demonstrated that the esp gene is a marker of the high prevalence of Enterococcus strains resistant to vancomycin in hospitalized patients,30 but according to the study of Woodford et al.,31 Sauer et al.,32 and Jahangiri et al.12 the esp gene also was identified in sensitive-vancomycin strains. In some studies, the prevalence of gene esp is reversed among VREfm and VSEfm strains. Whereas in other studies, the prevalence of gene esp was identical in VREfm and VSEfm strains. In accordance with the majority of these investigations, in the present study, the gene esp also was identified in a large number of VREfm strains (61%) compared with in VSEfm (21%). Camargo et al.33 demonstrated that esp was restricted to VREfm (56%) and not found in VSEfm. Vankerckhoven et al.10 surveyed virulence genes in eight European hospitals and found higher prevalences of esp in the clinical VREfm isolates (77%). Worth et al.34 and Sharifi et al.13 also found a higher incidence of esp in 80.5% and 71.05% in the clinical VREfm isolates, respectively. In the study by Terkuran et al.35 the esp gene was detected in 15.7% of VREfm isolates and the hyl gene in 28.6% of VREfm strains, but these genes were identified in 2.9% of Enterococcus sensitive and intermediate strains. In the study by Sauer et al.32 the esp gene was identified in 62.9% and 46.3% of VRE and VSE strains, respectively. Also, in the study by Jahangiri et al.12 esp gene was identified in 82% of clinical VREfm isolates and 53% of VSEfm isolates. The esp and hyl genes have significantly a higher prevalence among ampicillin-resistant VREfm isolates (53.7%, 37.3%) than ampicillin-susceptible VREfm isolates (19.4%, 22.4%); which is similar to the results of other studies.3334353637 The high frequency of esp gene, which was shown in the present study and most of the analogous studies, could be due to the fact that strains containing this gene can obtain antibiotic resistant genes and antibiotic resistant bacteria have long term stability in the body.2327
In the present study, the asa1 gene, (which encodes aggregation substance), was found in high frequency among E. faecalis (97%) and E. faecium strains (100%). A high incidence of this gene in E. faecalis was reported in previous studies. Results of studies on clinical E. faecium isolates are contradictory. In some studies, asa1 was not found in E. faecium but in contrast, in some studies this gene was detected in lower frequency and in our study and some other studies this gene was identified in higher prevalence among E. faecium isolates (Comerlato et al.38 and Kowalska-Krochmal et al.39), were detected it among 5%, 65% of VREfm and 2.7%, 60% of VREfs strains, respectively. Jahangiri et al.12 were not found asa1 gene in either 49 of VREfm strains or 17 of VSEfm strains. Sharifi et al.13 were detected asa1 gene in 80% of VREfs and 7.89% of VREfm strains. Hällgren et al.24 also were reported prevalence of asa1 in 79% of E. faecium strains. In studies by Huyckl & Gilmore, the asa1 gene was detected in 100% of blood isolates and 32% of non-blood isolates of Enterococcus. In studies by Elsner et al, Eaton & Gasson, and Archimbaud et al, the asa1 gene was detected in 40%-78% of clinical isolates of Enterococci.24 Hyaluronidase, coded by the chromosomal gene hyl, that influence the hyaluronic acid (hyaluronate, HA).35 Hyaluronidase in Enterococcus, indicates some homology to the hyaluronidases in other bacteria such as, Streptococcus pyogenes, Staphylococcus aureus and Streptococcus pneumoniae.9 We found the hyl gene among 49.3% of VREfm and 22.4% of VSEfm isolates, which is in accordance to findings of Rice et al.40 who detected the hyl gene among 71% of the United Kingdom VREfm isolates. But it was in contrast to the study by Jahangiri et al.12 which detected hyl gene among 80% of VSEfm and in 28.5% of VREfm isolates.
The reasons for the diversity in frequencies of hyl and asa1 can be as follows; Entrococcus strains are genetically different from each other based on their geographical origins. Moreover, the media which were used as sampling sources were varied between studies. In other words, some studies took their samples from blood, while others used urine, foods, or sewage.38263541 As demonstrated by Billström et al.19 and Wardal et al.42 the esp and hyl genes are linked with ampicillin and ciprofloxacin-resistant enterococci, particularly in CC17 which is an especially virulent, hospital adapted clone found globally. In the present study, it was shown that the esp virulence gene has a significant association with ciprofloxacin (p=0.001), erythromycin (p=0.001) and tetracycline (p=0.001) susceptibility patterns in E. faecium and with chloramphenicol (p=0.001) in E. faecalis strains; the hyl with teicoplanin (p=0.001) and vancomycin (p=0.001) in E. faecium strains; and also asa1 with vancomycin (p=0.001) in E. faecium and with ampicillin (p= 0.001) and chloramphenicol (p=0.001) in E. faecalis strains (Table 5).
The correlation between infective genes and antibiotic resistance in Enterococcus may vary from country to country. Based on the study by Hanna Billström et al.19 a significant relationship between imipenem, ampicillin, and ciprofloxacin resistance pattern and the espfm gene was found. Resistance to ciprofloxacin, imipenem, ampicillin antibiotics and the prevalence of the esp and hyl genes in these isolates were reported 90%, 80%, 77%, 56% and 4%, respectively. In the Study by Baylan et al.43 (Turkey, 2008) on E. faecalis strains, a significant association between the carriage asa1 gene and ciprofloxacin, norfloxacin and levofloxacin resistance pattern and between the esp gene and doxycycline resistance pattern were observed; in addition, a significant association between the hyl gene and nitrofurantoin resistance pattern in E. faecium strains was indicated. In the other study by Jankoska et al.41 there was no significant relationship between virulence genes and antibiotic resistance patterns; the esp gene was detected in 76% of isolates and all strains were susceptible to vancomycin and nitrofurantoin, 24%, 34%, and 28% of strains were resistant to ampicillin, ciprofloxacin and ceftriaxone, respectively.
According to the results of studies by Jahangiri et al.12 and Duprè,44 it was found that most of esp-positive isolates were resistant to more than 3 antibiotics. Lund et al.45 demonstrated that the existence of a strong correlation between the carriage of esp gene and antimicrobial resistance could be due to the higher conjugation frequencies in strains carrying the esp gene compared with strains lacking this gene. Sharifi et al.13 showed that E. faecium strains carrying the esp gene were resistant to more than 90% of the tested antibiotics and 64% of them were resistant to vancomycin. Considering these results, it seems that the esp gene facilitates E. faecium isolates ability to acquire antibiotic resistance genes. Van Wamel et al.46 showed that the expression level of the esp gene vary constantly between E. faecium strains depending on growth conditions and it is associated with bothe the initial connection and biofilm formation. Due to increasing resistance rates of enterococci to most common antibiotics, strict infection control measures are required. Antibiotic susceptibility testing is recommended for all patients before treatment for rational antibiotic use. There is a significant relationship between virulence genes and antibiotic resistance patterns. The virulence factors involve conjugative transfer of antibiotic resistance genes among enterococci strains and other species especially as the transfer of vancomycin resistance to staphylococcus aureus strains may occur.
Figures and Tables
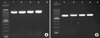 | FIG. 1PCR amplification of ddl E. faecalis, ddl E. faecium genes. (A) PCR products ddl E. faecalis gene (941 bp). (B) PCR products ddl E. faecium genes (658 bp). L: molecular size marker 100 bp, 1: positive control, 2-4: samples, 5: negative control. |
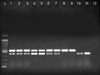 | FIG. 2PCR amplification of esp, asa1 genes. PCR products esp, asa1 virulence genes is 510 bp and 375 bp respectively. L: molecular size marker 100 bp, 1: positive control, 2-11: samples, 12: negative control. |
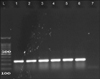 | FIG. 3PCR amplification of hyl gene. PCR product hyl virulence gene is 275 bp. L: molecular size marker 100 bp, 1: positive control, 2-6: samples, 7: negative control. |
TABLE 2
Distribution of E. faecalis and E. faecium strains isolated from Hamadan hospitals based on the sample source

ACKNOWLEDGEMENTS
The authors are grateful to thank from Prof. Duygu Percin for reading, editing and giving some valuable comments and Vice chancellor of Hamadan University for supporting funding.
References
1. Nateghian AR, Ghasemi Ahari SM, Lahouti Harahdashti A, Navidnia M, Mehrazma M. Prevalence of vancomycin-resistant enterococci colonization, and susceptibility to linezolid in pediatric intensive care units of a referral pediatric center in Tehran, Iran. Arch Pediatr Infect Dis. 2014; 2:e16970.

2. Fernandes SC, Dhanashree B. Drug resistance & virulence determinants in clinical isolates of Enterococcus species. Indian J Med Res. 2013; 137:981–985.
3. Mira MU, Deana M, Xora J, Vera G, Biljana M, Biljana R. Prevalence of different enterococcal species isolated from blood and their susceptibility to antimicrobial drugs in Vojvodina, Serbia, 2011-2013. Afr J Microbiol Res. 2014; 8:819–824.

4. Samadi Kafil H, Mohabati Mobarez A, Fourouzandeh Moghadam M. Multidrug resistant and most virulent Enterococcus faecium (strain 2653), isolated from hospitalized patient wound in Iran. Scholar J Med. 2012; 2:36–39.
5. Van Tyne D, Gilmore MS. Friend turned foe: evolution of enterococcal virulence and antibiotic resistance. Annu Rev Microbiol. 2014; 68:337–356.

6. Feizabadi MM, Sayadi S, Shokrzadeh L, Parvin M, Yadegarynia D. Increase in prevalence of vancomycin resistant isolates of Enterococcous faecium at Labbafinejad hospital. Iran J Clin Infect Dis. 2008; 3:73–77.
7. Medeiros AW, Pereira RI, Oliveira DV, Martins PD, d'Azevedo PA, Van der Sand S, et al. Molecular detection of virulence factors among food and clinical Enterococcus faecalis strains in South Brazil. Braz J Microbiol. 2014; 45:327–332.

8. Giridhara Upadhyaya PM, Umapathy BL, Ravikumar KL. Comparative study for the presence of enterococcal virulence factors gelatinase, hemolysin and biofilm among clinical and commensal isolates of enterococcus faecalis. J Lab Physicians. 2010; 2:100–104.

9. Biendo M, Adjidé C, Castelain S, Belmekki M, Rousseau F, Slama M, et al. Molecular characterization of glycopeptide-resistant enterococci from hospitals of the picardy region (france). Int J Microbiol. 2010; 2010:150464.

10. Vankerckhoven V, Huys G, Vancanneyt M, Snauwaert C, Swings J, Klare I, et al. Genotypic diversity, antimicrobial resistance, and virulence factors of human isolates and probiotic cultures constituting two intraspecific groups of Enterococcus faecium isolates. Appl Environ Microbiol. 2008; 74:4247–4255.

11. Camargo IL, Zanella RC, Gilmore MS, Darini AL. Virulence factors in vancomycin-resistant and vancomycin-susceptible Enterococcus faecalis from Brazil. Braz J Microbiol. 2008; 39:273–278.

12. Jahangiri S, Talebi M, Eslami G, Pourshafie MR. Prevalence of virulence factors and antibiotic resistance in vancomycin-resistant Enterococcus faecium isolated from sewage and clinical samples in Iran. Indian J Med Microbiol. 2010; 28:337–341.

13. Sharifi Y, Hasani A, Ghotaslou R, Varshochi M, Hasani A, Aghazadeh M, et al. Survey of Virulence Determinants among Vancomycin Resistant Enterococcus faecalis and Enterococcus faecium Isolated from Clinical Specimens of Hospitalized Patients of North west of Iran. Open Microbiol J. 2012; 6:34–39.

14. Manero A, Blanch AR. Identification of Enterococcus spp. with a biochemical key. Appl Environ Microbiol. 1999; 65:4425–4430.

15. Kariyama R, Mitsuhata R, Chow JW, Clewell DB, Kumon H. Simple and reliable multiplex PCR assay for surveillance isolates of vancomycin-resistant enterococci. J Clin Microbiol. 2000; 38:3092–3095.

16. Creti R, Imperi M, Bertuccini L, Fabretti F, Orefici G, Di Rosa R, et al. Survey for virulence determinants among Enterococcus faecalis isolated from different sources. J Med Microbiol. 2004; 53:13–20.

17. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility tests: approved standard - eleventh edition. CLSI document M02-A11. Wayne, PA: Clinical and Laboratory Standards Institute;2012.
18. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: twenty-Third informational supplement. CLSI document M100-S23. Wayne, PA: Clinical and Laboratory Standards Institute;2013.
19. Billström H, Lund B, Sullivan A, Nord CE. Virulence and antimicrobial resistance in clinical Enterococcus faecium. Int J Antimicrob Agents. 2008; 32:374–377.

20. Nowakiewicz A, Ziółkowska G, Zięba P, Trościańczyk A, Banach T, Kowalski C. Modified 16S-23S rRNA intergenic region restriction endonuclease analysis for species identification of Enterococcus strains isolated from pigs, compared with identification using classical methods and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Med Microbiol. 2015; 64:217–223.

21. Lavová M, Bezeková J, Čanigová M, Kročko M, Domig K. Species identification of Enterococci by biochemical tests and molecular-genetic methods. Potravinarstvo. 2014; 8:124–129.

22. Jurkovic D, Krizková L, Dusinský R, Belicová A, Sojka M, Krajcovic J, et al. Identification and characterization of enterococci from bryndza cheese. Lett Appl Microbiol. 2006; 42:553–559.

23. Sharifi Y, Hasani A, Ghotaslou R, Naghili B, Aghazadeh M, Milani M, et al. Virulence and antimicrobial resistance in enterococci isolated from urinary tract infections. Adv Pharm Bull. 2013; 3:197–201.
24. Hällgren A, Claesson C, Saeedi B, Monstein HJ, Hanberger H, Nilsson LE. Molecular detection of aggregation substance, enterococcal surface protein, and cytolysin genes and in vitro adhesion to urinary catheters of Enterococcus faecalis and E. faecium of clinical origin. Int J Med Microbiol. 2009; 299:323–332.

25. Eaton TJ, Gasson MJ. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl Environ Microbiol. 2001; 67:1628–1635.

26. Eaton TJ, Gasson MJ. A variant enterococcal surface protein Esp(fm) in Enterococcus faecium; distribution among food, commensal, medical, and environmental isolates. FEMS Microbiol Lett. 2002; 216:269–275.

27. Vankerckhoven V, Van Autgaerden T, Vael C, Lammens C, Chapelle S, Rossi R, et al. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J Clin Microbiol. 2004; 42:4473–4479.

28. Shankar V, Baghdayan AS, Huycke MM, Lindahl G, Gilmore MS. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect Immun. 1999; 67:193–200.

29. Channaiah LH, Subramanyam B, McKinney LJ, Zurek L. Stored-product insects carry antibiotic-resistant and potentially virulent enterococci. FEMS Microbiol Ecol. 2010; 74:464–471.

30. Willems RJ, Homan W, Top J, van Santen-Verheuvel M, Tribe D, Manzioros X, et al. Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet. 2001; 357:853–855.

31. Woodford N, Soltani M, Hardy KJ. Frequency of esp in Enterococcus faecium isolates. Lancet. 2001; 358:584.

32. Sauer P, Síla J, Vágnerová I. Virulence factors in vancomycin-susceptible and vancomycin-resistant enterococci in the University Hospital Olomouc. Klin Mikrobiol Infekc Lek. 2009; 15:44–47.
33. Camargo IL, Gilmore MS, Darini AL. Multilocus sequence typing and analysis of putative virulence factors in vancomycin-resistant and vancomycin-sensitive Enterococcus faecium isolates from Brazil. Clin Microbiol Infect. 2006; 12:1123–1130.

34. Worth LJ, Slavin MA, Vankerckhoven V, Goossens H, Grabsch EA, Thursky KA. Virulence determinants in vancomycin-resistant Enterococcus faecium vanB: clonal distribution, prevalence and significance of esp and hyl in Australian patients with haematological disorders. J Hosp Infect. 2008; 68:137–144.

35. Terkuran M, Erginkaya Z, Ünal E, Guran M, Kizilyildirim S, Gökce U, et al. The relationship between virulence factors and vancomycin resistance among Enterococci collected from food and human samples in Southern Turkey. Ankara Üniv Vet Fak Derg. 2014; 61:133–140.

36. Coque TM, Willems R, Cantón R, Del Campo R, Baquero F. High occurrence of esp among ampicillin-resistant and vancomycin-susceptible Enterococcus faecium clones from hospitalized patients. J Antimicrob Chemother. 2002; 50:1035–1038.

37. Leavis HL, Willems RJ, Top J, Spalburg E, Mascini EM, Fluit AC, et al. Epidemic and nonepidemic multidrug-resistant Enterococcus faecium. Emerg Infect Dis. 2003; 9:1108–1115.
38. Comerlato CB, Resende MC, Caierão J, d'Azevedo PA. Presence of virulence factors in Enterococcus faecalis and Enterococcus faecium susceptible and resistant to vancomycin. Mem Inst Oswaldo Cruz. 2013; 108:590–595.

39. Kowalska-Krochmal B, Dworniczek E, Dolna I, Bania J, Wałecka E, Seniuk A, et al. Resistance patterns and occurrence of virulence determinants among GRE strains in southwestern Poland. Adv Med Sci. 2011; 56:304–310.

40. Rice LB, Carias L, Rudin S, Vael C, Goossens H, Konstabel C, et al. A potential virulence gene, hylEfm, predominates in Enterococcus faecium of clinical origin. J Infect Dis. 2003; 187:508–512.

41. Jankoska G, Trajkovska-Dokic E, Panovski N, Popovska-Jovanovska K, Petrovska M. Virulence factors and antibiotic resistance in Enterococcus faecalis isolated from urine samples. Prilozi. 2008; 29:57–66.
42. Wardal E, Markowska K, Zabicka D, Wróblewska M, Giemza M, Mik E, et al. Molecular analysis of vanA outbreak of Enterococcus faecium in two Warsaw hospitals: the importance of mobile genetic elements. Biomed Res Int. 2014; 2014:575367.
43. Baylan O, Nazik H, Bektöre B, Citil BE, Turan D, Ongen B, et al. The relationship between antibiotic resistance and virulence factors in urinary Enterococcus isolates. Mikrobiyol Bul. 2011; 45:430–445.
44. Duprè I, Zanetti S, Schito AM, Fadda G, Sechi LA. Incidence of virulence determinants in clinical Enterococcus faecium and Enterococcus faecalis isolates collected in Sardinia (Italy). J Med Microbiol. 2003; 52:491–498.





 PDF
PDF ePub
ePub Citation
Citation Print
Print






 XML Download
XML Download