Abstract
Circulating tumor cells (CTCs) are tumor cells that are separated from the primary site or metastatic lesion and disseminate in blood circulation. CTCs are considered to be part of the long process of cancer metastasis. As a 'liquid biopsy', CTC molecular examination and investigation of single cancer cells create an important opportunity for providing an understanding of cancer biology and the process of metastasis. In the last decade, we have seen dramatic development in defining the role of CTCs in lung cancer in terms of diagnosis, genomic alteration determination, treatment response and, finally, prognosis prediction. The aims of this review are to understand the basic biology and to review methods of detection of CTCs that apply to the various types of solid tumor. Furthermore, we explored clinical applications, including treatment monitoring to anticipate therapy resistance as well as biomarker analysis, in the context of lung cancer. We also explored the potential use of cell-free circulating tumor DNA (ctDNA) in the genomic alteration analysis of lung cancer.
Lung cancer remains the main cause of mortality among patients with cancer worldwide.12 Among American patients with cancer, it is estimated that lung cancer is responsible for the mortality of 28% male and 26% female patients.3 Efforts have been made to detect lung cancer as early as possible by methods such as chest computerized tomography (CT) scanning,456 sputum cytology analysis,78 biomarker analysis,910 circulating tumor cell (CTC) detection, 11 and circulating cell-free tumor DNA (ctDNA) detection.1213 An average solid tumor may release an estimated million cells per day into the bloodstream and although most dispersed cancer cells do not survive, the cells that do survive clearly pose an existential threat to the host organism.14
Detection, monitoring, and molecular investigation of CTCs and ctDNA offer a meaningful and noninvasive way for the detection of the early phase of the disease, prognosis prediction and assessing therapeutic response in patients with lung cancer.1516 As a so-called 'liquid biopsy', single tumor cell analysis and ctDNA analysis hold significant potential in providing understanding into lung cancer biology and the cancer spreading process.17 Until now CTC characterization and enumeration has been extensively studied in several cancers such as colorectal carcinoma,1819 prostate cancer,2021 breast cancer,2223 ovarian cancer,2425 head and neck carcinoma,2627 brain cancer,282930 gastric cancer,3132 hepatocellular carcinoma,333435 renal cell carcinoma,36 mesothelioma,3738 and lung cancer.3940 In general, the objectives of studies on CTCs in solid tumor include (a) risk prediction for metastatic progression and relapse , (b) staging determination and real-time observation of response to therapies, (c) determination of therapeutic targets and mechanisms of resistance, and (d) understanding metastasis process in solid tumor patients.41
Circulating ctDNA can be identified in lung cancer patients with the polymerase chain reaction (PCR) assay and high plasma DNA levels could also identify high-risk persons for lung cancer screening.42 Moreover, ctDNA in patients with lung cancer displays genetic and epigenetic differences common for tumors including oncogene activation, chromosome loss, and tumor-suppressor gene silencing by methylation.43 Studies have shown that ctDNA levels were associated with tumor grade, tumor stage, lymph node spreading, the number of metastatic sites, tumor response to treatment and, finally, survival in patients with non-small cell lung cancer (NSCLC).44
The purpose of this review is to discuss the basic concept and methods of detection of CTCs and ctDNA in solid tumors generally. We also aimed to see how these advancements of knowledge have been applied in assessing patients with lung cancer, both small NSCLC as well as small cell lung cancer, based on accumulated evidence. A review of the published peer-reviewed reports in the Medline database up to October 2015 was performed. The terms used to gather relevant original articles, reports, and reviews were: circulating tumor cells, circulating cell-free tumor DNA, lung cancer, and epithelial-mesenchymal transition. A narrative review format was used to present the information collected.
CTCs, with a spectrum from one cell to aggregates consisting of 2-5 cells, are tumor cells that detach from either a primary lesion or a metastatic site and spread in the peripheral blood as the cellular seed of metastasis (Fig. 1).4546 1 gram of tumor tissue, about 1 million tumor cells, can disseminate daily into the blood stream.47 CTCs in blood circulation derive from solid tumors. They are involved in the metastatic spread to distant organs that leads to the formation of secondary sites of the disease.48 Meanwhile, circulating tumor microemboli (CTM) is a tumor cell aggregate that circulates in the blood.49 Another term is disseminating tumor cells (DTCs), which are defined as a settlement of CTCs in secondary organs and may stay in a quiescence state or may cause an observable metastasis.50 CTCs and DTCs in blood and bone marrow are considered as potential metastases-inducing cells.51 At least one CTC per 3 ml of blood was reported in 17 (65%) and one CTM per 3 ml of blood in 15 (58%) of 26 NSCLC cases.52
Cell-free circulating tumor DNA (ctDNA) may come from primary or metastatic tumor deposits and is enriched for detection of metastatic precursors.53 ctDNA genotyping has some advantages over CTCs because (1) CTCs must be separated from the much more abundant hematologic cells in the blood requiring comprehensive laboratory facilities to obtain a viable population of CTCs for study; (2) CTCs in circulation encounter substantial apoptosis and fragility leading to variability between different CTC assays; (3) most of the ctDNA genotyping methods require a minimum of special handling and do not depend on special equipment, and lastly; (4) ctDNA could be processed at the same time with plasma DNA from normal cells, which always exists in the bloodstream.54
CTC takes part in the long process of tumor metastasis, it is believed that metastasis is started by a sub-group of CTC seen in the blood of patients.55 Metastasis consists of the two steps: The first steps is the displacement of a cancer cell to a distant site, while the second step is related to the capacity of the cancer cell to generate a metastatic foci at that distant organ.56 To detach from the primary tumor, cancer cells needs to undergo a cellular process known as the epithelial-mesenchymal transition (EMT).57 EMT allows the tumor cells to gain motility and migratory capacity which results in penetration into bloodstream and circulation as CTC.58
The release of DNA into circulation is a common phenomenon in cancer patients due to apoptotic and necrotic processes characteristic of tumor cells.59 Methylation, mutations, microsatellite alterations, DNA integrity, and viral DNA can be studied in the ctDNA, as well as contributing factors of tumor DNA released into circulation which are tumor burden and tumor cell proliferation (Fig. 2).60 Cell-free DNA found in lung cancer patients originates primarily from tumor development and necrotic malignant cells that have been enveloped by macrophages rather than the chronic inflammatory response.6162
CTC identification, enumeration, and molecular analysis are very difficult because CTCs occur typically with a frequency 1 per 1067 leukocytes and the amount of available sample is very limited.6364 Many methods have been tested to detect and characterize CTCs in lung cancer patients.65 Most of the current strategies to enumerate CTC are generally based on CTC markers and size. In the surface marker-based strategy, CTC detection is based on epithelial cell adhesion molecule (EpCAM) and keratin markers.66 One of the methods that are commonly used to enumerate CTC is CellSearch® CTC Test which has been shown to consistently capture clinically important CTCs.67 So far, only the CellSearch® system has been approved by the FDA for CTCs detection of epithelial origin in whole blood and to assess prognosis.68 CellSearch® isolates CTCs by using the EpCAM-based enrichment technique.69 The nanotechnology approach in CTCs detection has also been studied. Gazouli and colleagues demonstrated a sensitive assay that combines magnetic beads, coupled with EpCAM and CK19 antibodies, and quantum dots (QDs) fluorescence detection for the detection of as little as 10 CTCs per ml blood samples.70 However, because CTCs of lung cancer often present non-epithelial characteristics and lose their epithelial markers during the EMT process, this detection method can become ineffective and result in poor sensitivity.5171 In breast cancer, loss of EpCAM is accompanied by the upregulation of caveolin1 (CAV1) in CTC. CTC detection devices using CAV1-EpCAM conjugated beads give significantly improved throughput compared to EpCAM only.72 However, there has been no reporting about the use of CAV1-EpCAM-based CTC detection in lung cancer.
To address the limitation of EpCAM-based strategies, efforts to classify CTCs based on their EMT markers and to elucidate the heterogeneity of CTC populations have been made.73 Recently, the CanPatrol CTC enrichment method was used to detect CTCs using EMT markers in various types of malignancies.74 This technique might offer a solution because EpCAM-based enrichment methods have led to missed CTC detection that has lost their EpCAM expression due to EMT.75
As for the size-based CTC detection, most commonly, microfluidic chips are used, which involves filtration of micro-channels to isolate the larger CTCs from the other blood components.76 One study reported that an ultra-high-throughput spiral microfluidic biochip device could enrich CTC with a fast processing time (7.5 mL blood within 10 min), 100% detection rate (10/10) of blood samples collected from patients with advanced-stage metastatic lung and breast cancers, and was able to capture 20 to 135 CTCs/mL with high purity of 1 CTC out of every 30-100 white blood cells.77 Another sized-based CTC detection, microcavity array (MCA) system connected with a small device for CTC detection independent of EpCAM expression, has been reported to be successful in isolating CTC populations from patients who had been determined as CTC negative by the CellSearch system.78 The MCA system is a microfabricated nickel filter with a rectangular MCA (10(4) cavities/filter) and the shape and porosity of the MCA were customized to efficiently identify small tumor cells on the microcavities under low flow resistance while at the same time allowing other blood cells to pass through.79
Ligand-targeted PCR (LT-PCR) is a system where CTCs, that were enriched from erythrocytes and leukocytes, were marked with a conjugate of a tumor-specific ligand and a synthesized oligonucleotide followed by quantitative PCR analysis.80 LT-PCR provide quantification of CTC in NSCLC patients with greater sensitivity, about 80% sensitivity of stage I/II, 67% sensitivity of stage III, and 93% sensitivity of stage IV NSCLC.81 An assay that identified live cells using an adenoviral probe that detected elevated telomerase activity present in almost all cancer cells, but not in normal cells, could detect CTC in 65% of lung cancer patients.82 ISET (RareCell Diagnostics, Paris, France) is a technology that does not rely on the tumor marker expression, in which CTCs are captured by filtration independent of the tumor-related markers, because of their large size compared to circulating white blood cells.83 Filtration using the ISET system could detect larger numbers of CTCs, including epithelial marker negative cancer cells and isolate CTM and allow molecular analysis.84 The challenge is not limited to detection only but also culturing. If we could culture and expand the number CTCs ex vivo, it would allow us to examine genotype and phenotype of the tumor cells. In this regard, the Thermoresponsive NanoVelcro CTC purification system has been investigated for culture expansion of CTC from NSCLC.85 The Thermoresponsive NanoVelcro is the third generation of NanoVelcro in which agent-coated nanostructured substrates immobilized CTC while maintaining cells viability and molecular integrity.45
Calculation of ctDNA could assist in objective response assessments, identification of subclinical residual disease and provide a noninvasive approach for tumor genotyping.86 The quantity of plasma ctDNA can be determined using quantitative Real-Time PCR.87 It has been shown to reach the level of 12.8 ng/mL in lung cancer.12 Novel methods called the 'cancer personalized profiling by deep sequencing' (CAPP-Seq) for quantifying ctDNA can detect ctDNA in all the patients with stage II-IV and in half of the patients with stage I NSCLC.88
In the future, CTC applications will not be limited to enumeration. Daily characterization using a high-content approach that studies protein expression, morphometrics, genotype profiling and eventually prediction of the metastasis of the disease from the primary tumors to distant organs will become possible.89 Circulating cancer cells may be reliable markers in the diagnostic procedures of lung lesions.90 Even if without obvious tumor nodules, the CTCs can be captured in patients with COPD and non-detectable lung malignancy.91 Generally, the rate of CTC detection is positively correlated with NSCLC patients' overall stage and metastatic status.92
It has been demonstrated that CTC investigation through serial blood sampling could help provide personalized medicine for SCLC.93 Circulating tumor cells, prior to the initiation of chemotherapy, are valuable predictors of specific progression-free survival (PFS) in Stage III patients with SCLC.94 CTC enumerations could also be used for overall survival (OS) and PFS prediction. It has been shown that patients with no CTCs detected before treatment have a prolonged PFS and OS and show a greater percentage of stable disease compared to patients with 1-3 and >3 CTCs which impose a higher risk of having progressive disease.95 In terms of evaluation after treatment, patients with positive detection of CTCs after therapy show a poorer response to therapy.96
Real-time PCR and melting curve analysis have been used to find sensitizing EGFR mutations in blood cells enriched in CTC.97 Analysis of EGFR can also be performed in captured CTCs by immunofluorescence staining.98 EGFR mutation analysis on DNA recovered from CTCs showed high sensitivity, including identification of the T790M secondary mutation which confers drug resistance.99 Using ctDNA specimens, it was found that follows up to the EGFR mutations in the circulation allow for identification of the T790M mutation for up to 344 days before patients suffered from disease progression.100 Consistent with this finding, studies that quantitatively detected activating EGFR mutations and the resistant T790 secondary mutation showed a 72.7% detection rate of EGFR mutation in all lung cancer patient enrolled in the study. The detection rate of the T790M mutation among patients who experienced progressive disease after EGFR-TKI treatment was 43.5%. The BEAMing (beads, emulsion, amplification, and magnetics) method was used for detection.101
As for KRAS mutation, it was reported that the colorimetric membrane array analysis technique could identify an activated KRAS mutation from CTC in various cancers.102103 This finding was then followed by the establishment of the platform-weighted chemiluminescent membrane array (WCHMA) which is able to detect the KRAS mutation from minimally 3 CTCs/ml blood with a sensitivity of 93% and a specificity of 94%.104 In the WCHMA method, the best intensity value for each gene, for example, KRAS, is calculated by the linear value of the chemiluminescent emission and is more accurate than the color concentration readings in colorimetric membrane array method.105 However, recent evidence suggests that ctDNA was superior and preferable as type of specimen for KRAS mutation analysis in lung malignancies compared to CTC DNA because it could lead to greater mutation detection than CTCs.40
CTCs have also been used clinically to diagnose echinoderm microtubule associated proteins like 4-anaplastic lymphoma kinase (EML4-ALK) gene rearrangement in NSCLC to select patients who are eligible to receive ALK inhibitor.106 ALK rearrangement could be determined in CTCs of ALK-positive NSCLC patients by applying a filtration technique and filter adapted fluorescence in situ hybridization (FA-FISH).107 FA-FISH comprises a filter for detecting CTCs that have been previously enriched by blood filtration, followed by preparation of a filter spot for cells fixation and the cells analysis using the FISH assay.108 Moreover, ALK protein expression could also be observed in CTCs isolated from lung cancer patients by immunocytochemistry.109 FISH analysis provided sufficient positivity of EML4-ALK gene rearrangement detection from 10-1535 CTCs/mL and only required 7.5 mL of patient blood samples.110
Taken together, these advancements show that genetic modifications in solid cancers can be characterized by massively parallel sequencing of ctDNA shed from cancer cells into plasma. Repeated biopsies to investigate genomic dynamic changes as a consequence of therapy are not easy, invasive and may be confounded by the heterogeneity nature within the tumor.111 The potential of ctDNA as a replacement of metastasized tumor biopsy becomes more evident because, in some patients, mutation expression analysis from metastasis lesion biopsies failed because of insufficient biopsy sizes, but was successful in all plasma ctDNA samples.112 However, despite the promising application and their applicability for employing CTCs to diagnose genomic changes and monitor responses to therapies (ie, as a liquid biopsy) in lung cancer, these technologies have been limited by significant hurdles, such as complex systems that requires high-level laboratory capacity, contaminated blood cells, and undefined gold-standard method, and have not compiled momentum to add tissue-based diagnostics.113
CTCs are tumor cells that can be captured through a liquid biopsy from blood and can be genetically and phenotypically studied to provide important data for guiding cancer therapy. The clinical values of CTCs as a biomarker for early-phase cancer screening, diagnosis, the prediction of treatment response, prognosis, and stratification have been widely explored in recent years.114 It is expected that an understanding of CTC biology and its implications in their clinical application will help clinicians in the treatment of lung cancer. Despite high sensitivity and specificity, technological issues have limited the broad clinical utility of the method. It may need several years for CTC detection to become applicable in the clinic for the routine diagnosis of cancer. Obviously, further investigation is required to establish standardized techniques for sample collection, processing, and analysis.
Figures and Tables
FIG. 1
Circulating tumor cells (CTCs) and cell-free circulating tumor (ctDNA) originated from the primary tumor or metastatic lesion circulates in the blood. Reprinted with permission from Sysmex Inostics Corp.
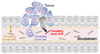
FIG. 2
Circulating cell-free tumor DNA exist in the blood in various forms, and could provide information such as mutations, DNA integrity, methylation, viral DNA, and microsatellite alterations. Reprinted by permission from Macmillan Publishers Ltd: Nature Reviews Cancer 11: 426-437, copyright (2011).

ACKNOWLEDGEMENTS
We would like to acknowledge Katrin Tomson for her support in proofreading and editing of this manuscript.
References
1. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015; 136:E359–E386.

2. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015; 65:87–108.

4. Kanodra NM, Silvestri GA, Tanner NT. Screening and early detection efforts in lung cancer. Cancer. 2015; 121:1347–1356.

5. Infante M, Cavuto S, Lutman FR, Passera E, Chiarenza M, Chiesa G, et al. Long-term follow-up results of the DANTE trial, a randomized study of lung cancer screening with spiral computed tomography. Am J Respir Crit Care Med. 2015; 191:1166–1175.

6. Shlomi D, Ben-Avi R, Balmor GR, Onn A, Peled N. Screening for lung cancer: time for large-scale screening by chest computed tomography. Eur Respir J. 2014; 44:217–238.

7. Sagawa M, Kobayashi T, Uotani C, Kibe Y, Tanaka M, Machida Y, et al. A survey about further work-up for cases with positive sputum cytology during lung cancer mass screening in Ishikawa Prefecture, Japan: a retrospective analysis about quality assurance of lung cancer screening. Jpn J Clin Oncol. 2015; 45:297–302.

8. Yu L, Shen J, Mannoor K, Guarnera M, Jiang F. Identification of ENO1 as a potential sputum biomarker for early-stage lung cancer by shotgun proteomics. Clin Lung Cancer. 2014; 15:372–378.e1.

9. Kim Y, Kim DH. CpG island hypermethylation as a biomarker for the early detection of lung cancer. Methods Mol Biol. 2015; 1238:141–171.

10. Hirales Casillas CE, Flores Fernández JM, Padilla Camberos E, Herrera López EJ, Leal Pacheco G, Martínez Velázquez M. Current status of circulating protein biomarkers to aid the early detection of lung cancer. Future Oncol. 2014; 10:1501–1513.

11. Yu N, Zhou J, Cui F, Tang X. Circulating tumor cells in lung cancer: detection methods and clinical applications. Lung. 2015; 193:157–171.

12. Paci M, Maramotti S, Bellesia E, Formisano D, Albertazzi L, Ricchetti T, et al. Circulating plasma DNA as diagnostic biomarker in non-small cell lung cancer. Lung Cancer. 2009; 64:92–97.

13. Sirera R, Bremnes RM, Cabrera A, Jantus-Lewintre E, Sanmartín E, Blasco A, et al. Circulating DNA is a useful prognostic factor in patients with advanced non-small cell lung cancer. J Thorac Oncol. 2011; 6:286–290.

14. Dong Y, Skelley AM, Merdek KD, Sprott KM, Jiang C, Pierceall WE, et al. Microfluidics and circulating tumor cells. J Mol Diagn. 2013; 15:149–157.

15. Han Y, Su C, Liu Z. Methods for detection of circulating cells in non-small cell lung cancer. Front Biosci (Landmark Ed). 2014; 19:896–903.

16. Karachaliou N, Mayo-de-Las-Casas C, Molina-Vila MA, Rosell R. Real-time liquid biopsies become a reality in cancer treatment. Ann Transl Med. 2015; 3:36.
17. Tognela A, Spring KJ, Becker T, Caixeiro NJ, Bray VJ, Yip PY, et al. Predictive and prognostic value of circulating tumor cell detection in lung cancer: a clinician's perspective. Crit Rev Oncol Hematol. 2015; 93:90–102.

18. Bork U, Rahbari NN, Schölch S, Reissfelder C, Kahlert C, Büchler MW, et al. Circulating tumour cells and outcome in non-metastatic colorectal cancer: a prospective study. Br J Cancer. 2015; 112:1306–1313.

19. Barbazán J, Muinelo-Romay L, Vieito M, Candamio S, Díaz-López A, Cano A, et al. A multimarker panel for circulating tumor cells detection predicts patient outcome and therapy response in metastatic colorectal cancer. Int J Cancer. 2014; 135:2633–2643.

20. Goldkorn A, Ely B, Tangen CM, Tai YC, Xu T, Li H, et al. Circulating tumor cell telomerase activity as a prognostic marker for overall survival in SWOG 0421: a phase III metastatic castration resistant prostate cancer trial. Int J Cancer. 2015; 136:1856–1862.

21. Thalgott M, Heck MM, Eiber M, Souvatzoglou M, Hatzichristodoulou G, Kehl V, et al. Circulating tumor cells versus objective response assessment predicting survival in metastatic castration-resistant prostate cancer patients treated with docetaxel chemotherapy. J Cancer Res Clin Oncol. 2015; 141:1457–1464.

22. Helissey C, Berger F, Cottu P, Diéras V, Mignot L, Servois V, et al. Circulating tumor cell thresholds and survival scores in advanced metastatic breast cancer: the observational step of the CirCe01 phase III trial. Cancer Lett. 2015; 360:213–218.

23. Beije N, Jager A, Sleijfer S. Circulating tumor cell enumeration by the CellSearch system: the clinician's guide to breast cancer treatment? Cancer Treat Rev. 2015; 41:144–150.

24. Pearl ML, Dong H, Tulley S, Zhao Q, Golightly M, Zucker S, et al. Treatment monitoring of patients with epithelial ovarian cancer using invasive circulating tumor cells (iCTCs). Gynecol Oncol. 2015; 137:229–238.

25. Kolostova K, Spicka J, Matkowski R, Bobek V. Isolation, primary culture, morphological and molecular characterization of circulating tumor cells in gynecological cancers. Am J Transl Res. 2015; 7:1203–1213.
26. Weller P, Nel I, Hassenkamp P, Gauler T, Schlueter A, Lang S, et al. Detection of circulating tumor cell subpopulations in patients with head and neck squamous cell carcinoma (HNSCC). PLoS One. 2014; 9:e113706.

27. Kulasinghe A, Perry C, Jovanovic L, Nelson C, Punyadeera C. Circulating tumour cells in metastatic head and neck cancers. Int J Cancer. 2015; 136:2515–2523.

28. Adamczyk LA, Williams H, Frankow A, Ellis HP, Haynes HR, Perks C, et al. Current understanding of circulating tumor cells - potential value in malignancies of the central nervous system. Front Neurol. 2015; 6:174.

29. Lombard A, Goffart N, Rogister B. Glioblastoma circulating cells: reality, trap or illusion? Stem Cells Int. 2015; 2015:182985.

30. Sullivan JP, Nahed BV, Madden MW, Oliveira SM, Springer S, Bhere D, et al. Brain tumor cells in circulation are enriched for mesenchymal gene expression. Cancer Discov. 2014; 4:1299–1309.

31. Lee SJ, Lee J, Kim ST, Park SH, Park JO, Park YS, et al. Circulating tumor cells are predictive of poor response to chemotherapy in metastatic gastric cancer. Int J Biol Markers. 2015; 30:e382–e386.

32. Kolostova K, Matkowski R, Gürlich R, Grabowski K, Soter K, Lischke R, et al. Detection and cultivation of circulating tumor cells in gastric cancer. Cytotechnology. 2016; 68:1095–1102.

33. Liu HY, Qian HH, Zhang XF, Li J, Yang X, Sun B, et al. Improved method increases sensitivity for circulating hepatocellular carcinoma cells. World J Gastroenterol. 2015; 21:2918–2925.

34. Fan JL, Yang YF, Yuan CH, Chen H, Wang FB. Circulating tumor cells for predicting the pognostic of patients with hepatocellular varcinoma: a meta analysis. Cell Physiol Biochem. 2015; 37:629–640.

35. Nel I, David P, Gerken GG, Schlaak JF, Hoffmann AC. Role of circulating tumor cells and cancer stem cells in hepatocellular carcinoma. Hepatol Int. 2014; 8:321–329.

36. Gradilone A, Iacovelli R, Cortesi E, Raimondi C, Gianni W, Nicolazzo C, et al. Circulating tumor cells and "suspicious objects" evaluated through CellSearch® in metastatic renal cell carcinoma. Anticancer Res. 2011; 31:4219–4221.
37. Raphael J, Massard C, Gong IY, Farace F, Margery J, Billiot F, et al. Detection of circulating tumour cells in peripheral blood of patients with malignant pleural mesothelioma. Cancer Biomark. 2015; 15:151–156.

38. Bobek V, Kacprzak G, Rzechonek A, Kolostova K. Detection and cultivation of circulating tumor cells in malignant pleural mesothelioma. Anticancer Res. 2014; 34:2565–2569.
39. Myung JH, Roengvoraphoj M, Tam KA, Ma T, Memoli VA, Dmitrovsky E, et al. Effective capture of circulating tumor cells from a transgenic mouse lung cancer model using dendrimer surfaces immobilized with anti-EGFR. Anal Chem. 2015; 87:10096–10102.

40. Freidin MB, Freydina DV, Leung M, Montero Fernandez A, Nicholson AG, Lim E. Circulating tumor DNA outperforms circulating tumor cells for KRAS mutation detection in thoracic malignancies. Clin Chem. 2015; 61:1299–1304.

41. Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem. 2013; 59:110–118.

42. Sozzi G, Conte D, Leon M, Ciricione R, Roz L, Ratcliffe C, et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J Clin Oncol. 2003; 21:3902–3908.

44. Nie K, Jia Y, Zhang X. Cell-free circulating tumor DNA in plasma/serum of non-small cell lung cancer. Tumour Biol. 2015; 36:7–19.

45. Lin M, Chen JF, Lu YT, Zhang Y, Song J, Hou S, et al. Nanostructure embedded microchips for detection, isolation, and characterization of circulating tumor cells. Acc Chem Res. 2014; 47:2941–2950.

46. King MR, Phillips KG, Mitrugno A, Lee TR, de Guillebon AM, Chandrasekaran S, et al. A physical sciences network characterization of circulating tumor cell aggregate transport. Am J Physiol Cell Physiol. 2015; 308:C792–C802.

47. Chang YS, di Tomaso E, McDonald DM, Jones R, Jain RK, Munn LL. Mosaic blood vessels in tumors: frequency of cancer cells in contact with flowing blood. Proc Natl Acad Sci U S A. 2000; 97:14608–14613.

48. Yap TA, Lorente D, Omlin A, Olmos D, de Bono JS. Circulating tumor cells: a multifunctional biomarker. Clin Cancer Res. 2014; 20:2553–2568.

49. Carlsson A, Nair VS, Luttgen MS, Keu KV, Horng G, Vasanawala M, et al. Circulating tumor microemboli diagnostics for patients with non-small-cell lung cancer. J Thorac Oncol. 2014; 9:1111–1119.

50. Joosse SA, Gorges TM, Pantel K. Biology, detection, and clinical implications of circulating tumor cells. EMBO Mol Med. 2014; 7:1–11.

51. Gires O, Stoecklein NH. Dynamic EpCAM expression on circulating and disseminating tumor cells: causes and consequences. Cell Mol Life Sci. 2014; 71:4393–4402.

52. Mascalchi M, Falchini M, Maddau C, Salvianti F, Nistri M, Bertelli E, et al. Prevalence and number of circulating tumour cells and microemboli at diagnosis of advanced NSCLC. J Cancer Res Clin Oncol. 2016; 142:195–200.

53. Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014; 4:650–661.

54. Huang WL, Wei F, Wong DT, Lin CC, Su WC. The emergent landscape of detecting EGFR mutations using circulating tumor DNA in lung cancer. Biomed Res Int. 2015; 2015:340732.

55. Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V, et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol. 2013; 31:539–544.

57. Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013; 27:2192–2206.

58. Ogden A, Rida PC, Aneja R. Heading off with the herd: how cancer cells might maneuver supernumerary centrosomes for directional migration. Cancer Metastasis Rev. 2013; 32:269–287.

59. Ulivi P, Silvestrini R. Role of quantitative and qualitative characteristics of free circulating DNA in the management of patients with non-small cell lung cancer. Cell Oncol (Dordr). 2013; 36:439–448.

60. Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011; 11:426–437.

61. Szpechcinski A, Chorostowska-Wynimko J, Struniawski R, Kupis W, Rudzinski P, Langfort R, et al. Cell-free DNA levels in plasma of patients with non-small-cell lung cancer and inflammatory lung disease. Br J Cancer. 2015; 113:476–483.

62. Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem. 2015; 61:112–123.

63. Lianidou ES, Strati A, Markou A. Circulating tumor cells as promising novel biomarkers in solid cancers. Crit Rev Clin Lab Sci. 2014; 51:160–171.

64. McInnes LM, Jacobson N, Redfern A, Dowling A, Thompson EW, Saunders CM. Clinical implications of circulating tumor cells of breast cancer patients: role of epithelial-mesenchymal plasticity. Front Oncol. 2015; 5:42.

65. Hofman V, Ilie M, Long E, Guibert N, Selva E, Washetine K, et al. Detection of circulating tumor cells from lung cancer patients in the era of targeted therapy: promises, drawbacks and pitfalls. Curr Mol Med. 2014; 14:440–456.

66. Joosse SA, Pantel K. Biologic challenges in the detection of circulating tumor cells. Cancer Res. 2013; 73:8–11.

67. Adams DL, Stefansson S, Haudenschild C, Martin SS, Charpentier M, Chumsri S, et al. Cytometric characterization of circulating tumor cells captured by microfiltration and their correlation to the CellSearch(®) CTC test. Cytometry A. 2015; 87:137–144.

68. Truini A, Alama A, Dal Bello MG, Coco S, Vanni I, Rijavec E, et al. Clinical applications of circulating tumor cells in lung cancer patients by cellsearch system. Front Oncol. 2014; 4:242.

69. Schulze K, Gasch C, Staufer K, Nashan B, Lohse AW, Pantel K, et al. Presence of EpCAM-positive circulating tumor cells as biomarker for systemic disease strongly correlates to survival in patients with hepatocellular carcinoma. Int J Cancer. 2013; 133:2165–2171.

70. Gazouli M, Lyberopoulou A, Pericleous P, Rizos S, Aravantinos G, Nikiteas N, et al. Development of a quantum-dot-labelled magnetic immunoassay method for circulating colorectal cancer cell detection. World J Gastroenterol. 2012; 18:4419–4426.

71. Young R, Pailler E, Billiot F, Drusch F, Barthelemy A, Oulhen M, et al. Circulating tumor cells in lung cancer. Acta Cytol. 2012; 56:655–660.

72. Kim YJ, Koo GB, Lee JY, Moon HS, Kim DG, Lee DG, et al. A microchip filter device incorporating slit arrays and 3-D flow for detection of circulating tumor cells using CAV1-EpCAM conjugated microbeads. Biomaterials. 2014; 35:7501–7510.

73. Mohamadi RM, Besant JD, Mepham A, Green B, Mahmoudian L, Gibbs T, et al. Nanoparticle-mediated binning and profiling of heterogeneous circulating tumor cell subpopulations. Angew Chem Int Ed Engl. 2015; 54:139–143.

74. Wu S, Liu S, Liu Z, Huang J, Pu X, Li J, et al. Classification of circulating tumor cells by epithelial-mesenchymal transition markers. PLoS One. 2015; 10:e0123976.

75. Gorges TM, Tinhofer I, Drosch M, Röse L, Zollner TM, Krahn T, et al. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer. 2012; 12:178.

76. Huang T, Jia CP, Jun-Yang , Sun WJ, Wang WT, Zhang HL, et al. Highly sensitive enumeration of circulating tumor cells in lung cancer patients using a size-based filtration microfluidic chip. Biosens Bioelectron. 2014; 51:213–218.

77. Warkiani ME, Khoo BL, Tan DS, Bhagat AA, Lim WT, Yap YS, et al. An ultra-high-throughput spiral microfluidic biochip for the enrichment of circulating tumor cells. Analyst. 2014; 139:3245–3255.

78. Hosokawa M, Kenmotsu H, Koh Y, Yoshino T, Yoshikawa T, Naito T, et al. Size-based isolation of circulating tumor cells in lung cancer patients using a microcavity array system. PLoS One. 2013; 8:e67466.

79. Hosokawa M, Yoshikawa T, Negishi R, Yoshino T, Koh Y, Kenmotsu H, et al. Microcavity array system for size-based enrichment of circulating tumor cells from the blood of patients with small-cell lung cancer. Anal Chem. 2013; 85:5692–5698.

80. Chen X, Zhou F, Li X, Yang G, Zhang L, Ren S, et al. Folate receptor-positive circulating tumor cell detected by LT- PCR-based method as a diagnostic biomarker for non-small-cell lung cancer. J Thorac Oncol. 2015; 10:1163–1171.

81. Lou J, Ben S, Yang G, Liang X, Wang X, Ni S, et al. Quantification of rare circulating tumor cells in non-small cell lung cancer by ligand-targeted PCR. PLoS One. 2013; 8:e80458.

82. Dorsey JF, Kao GD, MacArthur KM, Ju M, Steinmetz D, Wileyto EP, et al. Tracking viable circulating tumor cells (CTCs) in the peripheral blood of non-small cell lung cancer (NSCLC) patients undergoing definitive radiation therapy: pilot study results. Cancer. 2015; 121:139–149.

83. Ma YC, Wang L, Yu FL. Recent advances and prospects in the isolation by size of epithelial tumor cells (ISET) methodology. Technol Cancer Res Treat. 2013; 12:295–309.

84. Krebs MG, Hou JM, Sloane R, Lancashire L, Priest L, Nonaka D, et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J Thorac Oncol. 2012; 7:306–315.

85. Ke Z, Lin M, Chen JF, Choi JS, Zhang Y, Fong A, et al. Programming thermoresponsiveness of NanoVelcro substrates enables effective purification of circulating tumor cells in lung cancer patients. ACS Nano. 2015; 9:62–70.

86. Bratman SV, Newman AM, Alizadeh AA, Diehn M. Potential clinical utility of ultrasensitive circulating tumor DNA detection with CAPP-Seq. Expert Rev Mol Diagn. 2015; 15:715–719.

87. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014; 6:224ra24.
88. Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014; 20:548–554.

89. Rodriguez-Lee M, Kuhn P, Webb DR. Advancing cancer patient care by integrating circulating tumor cell technology to understand the spatial and temporal dynamics of cancer. Drug Dev Res. 2014; 75:384–392.

90. Fiorelli A, Accardo M, Carelli E, Angioletti D, Santini M, Di Domenico M. Circulating tumor cells in diagnosing lung cancer: clinical and morphologic analysis. Ann Thorac Surg. 2015; 99:1899–1905.

91. Ilie M, Hofman V, Long-Mira E, Selva E, Vignaud JM, Padovani B, et al. "Sentinel" circulating tumor cells allow early diagnosis of lung cancer in patients with chronic obstructive pulmonary disease. PLoS One. 2014; 9:e111597.

92. Sheu CC, Yu YP, Tsai JR, Chang MY, Lin SR, Hwang JJ, et al. Development of a membrane array-based multimarker assay for detection of circulating cancer cells in patients with non-small cell lung cancer. Int J Cancer. 2006; 119:1419–1426.

93. Hodgkinson CL, Morrow CJ, Li Y, Metcalf RL, Rothwell DG, Trapani F, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med. 2014; 20:897–903.

94. Fu L, Liu F, Fu H, Liu L, Yuan S, Gao Y, et al. Circulating tumor cells correlate with recurrence in stage III small-cell lung cancer after systemic chemoradiotherapy and prophylactic cranial irradiation. Jpn J Clin Oncol. 2014; 44:948–955.

95. Bao H, Burke PA, Huang J, Chen X, Brohawn PZ, Yao Y, et al. Circulating tumor cells: application as a biomarker for molecular characterization and predictor of survival in an all-comer solid tumor phase I clinical study. PLoS One. 2013; 8:e58557.

96. Sher YP, Shih JY, Yang PC, Roffler SR, Chu YW, Wu CW, et al. Prognosis of non-small cell lung cancer patients by detecting circulating cancer cells in the peripheral blood with multiple marker genes. Clin Cancer Res. 2005; 11:173–179.
97. Breitenbuecher F, Hoffarth S, Worm K, Cortes-Incio D, Gauler TC, Köhler J, et al. Development of a highly sensitive and specific method for detection of circulating tumor cells harboring somatic mutations in non-small-cell lung cancer patients. PLoS One. 2014; 9:e85350.

98. Punnoose EA, Atwal SK, Spoerke JM, Savage H, Pandita A, Yeh RF, et al. Molecular biomarker analyses using circulating tumor cells. PLoS One. 2010; 5:e12517.

99. Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008; 359:366–377.

100. Sorensen BS, Wu L, Wei W, Tsai J, Weber B, Nexo E, et al. Monitoring of epidermal growth factor receptor tyrosine kinase inhibitor-sensitizing and resistance mutations in the plasma DNA of patients with advanced non-small cell lung cancer during treatment with erlotinib. Cancer. 2014; 120:3896–3901.

101. Taniguchi K, Uchida J, Nishino K, Kumagai T, Okuyama T, Okami J, et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res. 2011; 17:7808–7815.

102. Yang MJ, Chiu HH, Wang HM, Yen LC, Tsao DA, Hsiao CP, et al. Enhancing detection of circulating tumor cells with activating KRAS oncogene in patients with colorectal cancer by weighted chemiluminescent membrane array method. Ann Surg Oncol. 2010; 17:624–633.

103. Chen YF, Wang JY, Wu CH, Chen FM, Cheng TL, Lin SR. Detection of circulating cancer cells with K-ras oncogene using membrane array. Cancer Lett. 2005; 229:115–122.

104. Tsao DA, Yang MJ, Chang HJ, Yen LC, Chiu HH, Hsueh EJ, et al. A fast and convenient new technique to detect the therapeutic target, K-ras mutant, from peripheral blood in non-small cell lung cancer patients. Lung Cancer. 2010; 68:51–57.

105. Lin SR, Huang MY, Chang MS. A high-performance gene chip platform for detecting genetic markers from circulating tumor cells. Biomark Genom Med. 2013; 5:12–17.

106. Faugeroux V, Pailler E, Auger N, Taylor M, Farace F. Clinical utility of circulating tumor cells in ALK-positive non-small-cell lung cancer. Front Oncol. 2014; 4:281.

107. Pailler E, Adam J, Barthélémy A, Oulhen M, Auger N, Valent A, et al. Detection of circulating tumor cells harboring a unique ALK rearrangement in ALK-positive non-small-cell lung cancer. J Clin Oncol. 2013; 31:2273–2281.

108. Cho WC. Emerging techniques in molecular detection of circulating tumor cells. Expert Rev Mol Diagn. 2014; 14:131–134.

109. Ilie M, Long E, Butori C, Hofman V, Coelle C, Mauro V, et al. ALK-gene rearrangement: a comparative analysis on circulating tumour cells and tumour tissue from patients with lung adenocarcinoma. Ann Oncol. 2012; 23:2907–2913.

110. Khoo BL, Warkiani ME, Tan DS, Bhagat AA, Irwin D, Lau DP, et al. Clinical validation of an ultra high-throughput spiral microfluidics for the detection and enrichment of viable circulating tumor cells. PLoS One. 2014; 9:e99409.

111. Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013; 497:108–112.

112. Lebofsky R, Decraene C, Bernard V, Kamal M, Blin A, Leroy Q, et al. Circulating tumor DNA as a non-invasive substitute to metastasis biopsy for tumor genotyping and personalized medicine in a prospective trial across all tumor types. Mol Oncol. 2015; 9:783–790.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download