Abstract
Fibromyalgia (FM) affects 1% to 5% of the population, and approximately 90% of the affected individuals are women. FM patients experience impaired quality of life and the disorder places a considerable economic burden on the medical care system. With the recognition of FM as a major health problem, many recent studies have evaluated the pathophysiology of FM. Although the etiology of FM remains unknown, it is thought to involve some combination of genetic susceptibility and environmental exposure that triggers further alterations in gene expression. Because FM shows marked familial aggregation, most previous research has focused on genetic predisposition to FM and has revealed associations between genetic factors and the development of FM, including specific gene polymorphisms involved in the serotonergic, dopaminergic, and catecholaminergic pathways. The aim of this review was to discuss the current evidence regarding genetic factors that may play a role in the development and symptom severity of FM.
Go to : 
Fibromyalgia (FM) is estimated to affect from 1% to 5% of the general population, and most patients with FM are women.1 FM is characterized by chronic widespread pain and is commonly accompanied by a constellation of additional symptoms such as fatigue, sleep disturbance, joint stiffness, and depression. Although pain is the central symptom of FM, the severity of pain can vary between FM patients and pain may fluctuate in a specific patient at different times. Despite much debate regarding the classification and concept of FM in the past, FM is now recognized as a disorder.23
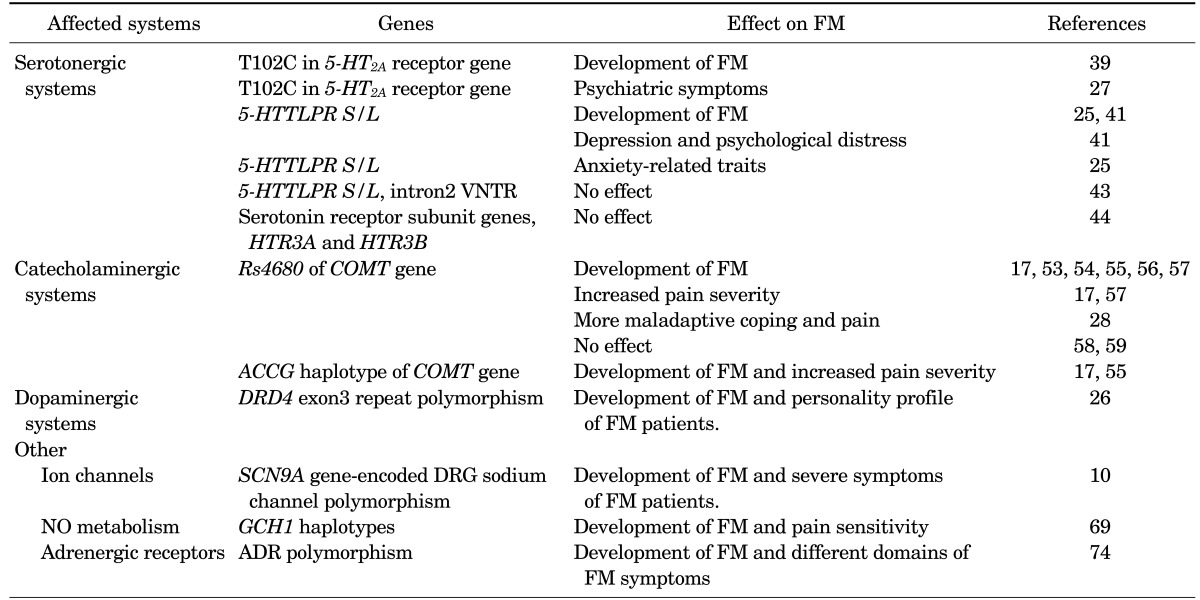
Patients with FM experience significantly impaired quality of life and their cost of care represents a significant economic burden.4 In fact, FM patients have similar health care resource requirements as patients with other common chronic diseases, such as diabetes mellitus and hypertension.5 For these reasons, FM is now recognized as a major health problem, and many recent studies have evaluated the pathophysiology of FM. Although the etiology of FM remains unknown, it is thought to involve the interaction of numerous factors, including psychological, genetic, neurobiological, and environmental factors.67 Owing to the occurrence of marked familial aggregation among FM sufferers, much previous research has focused on genetic predisposition to FM and has provided reasonable support for an association between genetic factors and the development of FM.8910 In this review, we explore the current evidence for potential genetic factors playing a role in the development and symptom severity of FM (Table 1).
Go to : 
Pain is known to have a genetic background in that it runs strongly in families in humans, which is due to pain-related genes that affect the expression or function of protein products in a way that affects the pain response.11 Individual differences in pain and analgesia can be explained by environmental as well as heritable factors.12 Pain genes have been evaluated in large familial studies using linkage analysis and in large cohorts of individuals with and without a specific disease by using association analysis.1113 Consequently, several genes have been shown to play major roles in pain sensitivity, including the genes encoding voltage-gated sodium-channels (Nav), GTP cyclohydrolase 1 (GCH1), mu-opioid receptors, and catechol-O-methyl transferase (COMT).14151617 In this context, familial aggregation and genome-wide association studies (GWAS) of chronic pain syndromes, including FM, have been performed to evaluate genetic susceptibility to disease.
Go to : 
Several studies have observed strong familial aggregation among FM patients. Pellegrino et al.18 investigated 17 families of patients with FM for evidence of inherited FM. In that study, 26 (52%) of the enrolled family members (parents and siblings) showed clinical evidence of FM, and an additional 11 individuals (22%) showed abnormal muscle consistency without tender points. However, the study participants were not diagnosed as having FM according to the 1990 American College of Rheumatology (ACR) criteria for the classification of FM.19
Buskila et al.20 evaluated the familial occurrence of FM among 58 offspring aged 5 to 46 years from 20 complete nuclear families with FM-diagnosed mothers. FM patients were diagnosed according to the ACR criteria,19 and 16 offspring (28%) were found to have FM. Offspring with and without FM did not differ significantly in terms of their exposure to environmental factors or other psychological or familial factors, such as anxiety, depression, global well-being, quality of life, or physical functioning. Thus, these results suggest that certain genetic factors are associated with the high familial occurrence of FM.
In another study, Buskila et al.21 studied female FM patients and their close relatives (parents, siblings, children, and husbands) to assess the familial aggregation of FM. The prevalence of FM among the blood relatives of patients with FM was 26%, compared to only 19% among unrelated family members (husbands). The occurrence of FM showed marked female preponderance: 41% of female relatives showed evidence of FM, compared to only 14% of male relatives. Additionally, pain sensitivity among young male and female relatives, which was measured by using mean tender point counts, was significantly higher than in controls. Moreover, adult relatives had considerably higher mean tender point counts than did controls. The authors concluded that the relatives of patients with FM demonstrated familial aggregation with respect to FM and were likely to have higher pain sensitivity than the general population.
Arnold et al.22 evaluated the familial aggregation of FM and pain sensitivity among 533 relatives of 78 probands with FM and compared these with 272 relatives of 40 probands with rheumatoid arthritis (RA). FM cases showed significant aggregation among the relatives of FM probands compared to those of RA probands (odds ratio [OR], 8.5; p=0.0002). The mean number of tender points was also significantly higher among the relatives of FM patients than among the relatives of patients with from RA.
Familial aggregation in FM does not exclude other possible etiologic factors in the development of FM. In fact, there is evidence that environmental factors also contribute to the development of FM.2324 However, familial aggregation does indicate that genetic factors are important to the etiology of FM. Subsequent investigations have demonstrated that FM is associated with genetic polymorphisms affecting the serotonergic,25 dopaminergic,26 and catecholaminergic systems.17 Furthermore, certain genetic polymorphisms influence not only the development of FM but also symptom severity.172728
Go to : 
GWAS are a recently developed research technique used to evaluate many common genetic variants in different individuals to determine whether a specific variant is associated with a given trait. As such, these studies are typically used to investigate disease heritability.29 GWAS usually focus on associations between single-nucleotide polymorphisms (SNPs) and traits, such as certain diseases that are associated with genetic background.2930 A linkage study has a similar aim to GWAS and is a recombinant technology used to map genetic loci by use of observations of related individuals.31
GWAS have been used to evaluate the association between FM and genetic susceptibility. Docampo et al.32 explored genetic susceptibility to FM through GWAS and array comparative genomic hybridization assessments of copy number variants. In that study, among 505,454 SNPs, none of the SNPs reached the genome-wide significance threshold. Nevertheless, the authors identified two associated variants, an SNP in the MYT1L gene and an intronic deletion in the NRXN3 gene, that suggested a possible role for the central nervous system in FM genetic susceptibility.32 Feng et al.33 also discovered possible candidate genes in FM by whole exome sequencing. They identified 2 nonsense mutations, W32X in C11orf40 and Q100X in ZNF77, associated with FM, one of which correlates with high plasma MCP-1 and IP-10 levels and the other with high plasma IL-12 levels. This result suggests a probable inflammatory basis of the development of the syndrome.
In addition, Arnold et al.34 performed a linkage scan, evaluating 341 markers in members of 116 families from the Fibromyalgia Family Study. They detected a signal on chromosome 17p11.2-q11.2, indicating a susceptibility locus for FM. However, subsequent research on this linkage study has not yet been reported.
The results of association and linkage studies have expanded our understanding of the genetic basis of FM and have suggested several genetic markers as susceptibility genes for FM. However, studies are not sufficient and thus these findings remain inconclusive. Therefore, further well-designed studies consisting of large study populations are needed.
Go to : 
Earlier research on genetic susceptibility in FM focused on the serotonergic pathway. Accumulating evidence supports the concept of disturbances in serotonin (5-HT) metabolism and neurotransmission in FM. This is based on several recent findings. Russell et al.3536 found that the level of serotonin metabolites was lower in serum and cerebrospinal fluid in FM patients. Similarly, Wolfe et al.37 showed that serum levels of 5-HT in FM patients were significantly lower than in controls. The serotonin transporter (5-HTT) is responsible for transport of 5-HT from synaptic spaces into presynaptic neurons, thus playing a key role in serotonergic neurotransmission.38 For these reasons, genetic variation in the serotonergic system has received attention as a possible genetic factor in the etiology of FM.
Bondy et al.39 investigated a silent polymorphism (T102C) in the 5-HT2A receptor gene in 168 FM patients and 115 healthy controls and found significant changes in genotype distribution among FM patients compared with the controls. Specifically, FM patients showed a decrease in the T/T genotype and an increase in both the T/C and C/C genotypes. In addition, pain severity was significantly higher in patients with the T/T genotype. Gürsoy et al.27 also investigated the role of the T102C polymorphism in the development of FM. In contrast to the findings of Bondy et al.,39 the T102C polymorphism of the 5-HT2A receptor gene did not differ significantly between patients and controls. However, the genotypes influenced symptom severity in FM patients. The T/T genotype was associated with psychiatric symptoms of FM, as measured by using the Symptom Checklist-90-Revised (SCL-90-R) test. In addition, patients with the T/T genotype demonstrated the lowest pain thresholds. Furthermore, a meta-analysis demonstrated that the 5-HT2A receptor T102C polymorphism conferred susceptibility to FM.40
Offenbaecher et al.41 analyzed the genotypes of the promoter region of the 5-HTT gene in patients with FM. The frequency of the S/S genotype of 5-HTT was higher among FM patients (31%) than among healthy controls (16%). Furthermore, FM patients with the S/S genotype exhibited severe symptoms of depression and psychological distress, as evidenced by higher scores on the Beck Depression Inventory (BDI) and the SCL-90-R compared to patients in the L/L and L/S groups. These results support the notion that altered serotonin metabolism is related to the development of FM and its symptoms.
Cohen et al.25 further evaluated the role of the 5-HTT promoter region (5-HTTLPR) polymorphism. Ninety-nine female FM patients from two Israeli ethnic groups (Jewish and Arab) were genotyped. In that study, each patient was assessed by using the Tridimensional Personality Questionnaire (TPQ), a self-reported instrument consisting of 100 yes/no questions. The results showed an association between FM and the 5-HTTLPR polymorphism in both Israeli ethnic groups. Furthermore, the 5-HTTLPR polymorphism was associated with the TPQ scores of the FM patients. There was a significant association between 5-HTTLPR genotype and the TPQ harm avoidance trait, which is consistent with the results of an earlier study by Lesch et al.42 that revealed that the short allele of this polymorphism is related to anxiety-related traits.
Gursoy43 reported that the 5-HTT gene polymorphism was not associated with FM patients who had normal psychiatric status. In that study, 53 mentally healthy FM patients and 60 healthy controls were included. However, the 5-HTTLPR polymorphism and the VNTR variant of the 5-HTT gene were not significantly different between the patients and controls. Additionally, Frank et al.44 showed no association between FM and either of the serotonin receptor subunit genes, HTR3A and HTR3B, in 48 patients with FM. Moreover, the association between the 5-HTTLPR polymorphism and FM was not observed in a meta-analysis.40
Thus, although the roles of specific genes involved in the serotonergic pathway in the pathophysiology of FM have not yet been confirmed, much evidence does support an association between genes involved in serotonin function and FM. In fact, studies have also shown that 5-HT3 receptor antagonists, such as ondansetron, granisetron, and tropisetron, are potential treatment options in the therapy of FM patients.454647 Because the serotonergic pathway influences psychiatric symptoms, such as depression, anxiety, and fatigue, which occur commonly in patients with FM,48 it is possible that serotonergic gene polymorphisms indirectly affect variable clinical features of FM.
Collectively, insufficient research is available to fully understand the role of serotonergic gene polymorphisms in FM. Further studies are needed to better understand the role of genes involved in serotonergic metabolism in the development of FM.
Major advances in our understanding of the pathophysiology of FM have resulted from the recognition of central sensitization. Central sensitization represents an enhancement in the function of neurons and circuits in central nociceptive pathways and thus results in pain hypersensitivity.4950 The development of central sensitization is related to the monoamine neurotransmitters,51 and COMT is a major enzyme that inactivates catecholamine neurotransmitters such as dopamine, epinephrine, and norepinephrine. Genetic mutations in the COMT gene can induce functional impairment in the COMT enzyme and alterations in COMT activity.51 Thus, polymorphism in the COMT gene has been suggested as a genetic factor associated with FM susceptibility and symptom severity.
The most widely investigated COMT gene polymorphism is the SNP rs4680. This SNP occurs in codon 158, resulting in a valine (Val GTG) to methionine (Met ATG) transition. Rs4680 can cause three possible genotypes and, as a result, alter the activity of the COMT enzyme. The H/H (GG; Val-158-Val) genotype gives rise to an effective enzyme, whereas the H/L (AG; Met-158-Val) genotype produces an intermediate-activity enzyme. The L/L (AA; Met-158-Met) genotype causes a defective enzyme that is incapable of effectively clearing catecholamines.52
To date, data regarding the association between rs4680 and susceptibility to FM are inconclusive. However, much evidence supports an association between this SNP and the development of FM. Matsuda et al.53 revealed an association between rs4680 and FM susceptibility in a Brazilian population. Similar associations in a variety of ethnic backgrounds have been described by several investigators: Gürsoy et al.54 in a Turkish population, Valgas-Alarcón et al.55 in a Spanish population, and Cohen et al.56 in an Israeli population. Furthermore, the role of rs4680 was not limited to increased FM susceptibility. Barbosa et al.57 showed that rs4680 was not only associated with the development of FM, but also with increased pain severity in Brazilian FM patients. In that study, the individual carriers of the homozygous mutant genotype for rs4680 presented a higher Fibromyalgia Impact Questionnaire (FIQ) score. In another study performed in a Spanish population, it was found that rs4680 played a key role in pain severity in FM patients.17 FM patients with the Met158Met genotype showed higher pain hypersensitivity to thermal and pressure stimuli than patients with the Val158Met genotype. Finan et al.28 also showed that rs4680 may affect FM pain symptoms in a 30-day electronic diary assessment in which patients recorded daily ratings of pain and two forms of maladaptive coping: pain catastrophizing and pain attention. The authors found that FM patients with the Met158Met genotype reported more maladaptive coping and pain when pain catastrophizing or pain attention was elevated, relative to those with the Val158Val genotype. This result suggests that genetic variation in the Val 158Met polymorphism may contribute to FM pain symptoms through pathways of pain-related cognition.
However, there is also evidence of no association between rs4680 and the development of FM.5859 In a recent meta-analysis, stratification by ethnicity failed to reveal an association between rs4680 and FM in European or Turkish populations.60 Using an unpublished dataset, we also investigated the association between rs4860 and FM susceptibility in a Korean population and found that rs4860 was not associated with FM susceptibility.
Although other COMT gene polymorphisms have been identified, including the SNPs rs6269, rs4633, and rs4818, the roles of these variants in FM are not yet fully understood and have been contradictory in some cases.175557 Thus, investigators have paid attention to the haplotype of the COMT gene rather than individual SNPs. The haplotype refers to the set of alleles of a group of closely linked individual SNPs that are usually inherited as a unit and is critical for evaluating the genetic basis of diseases.61 Certain haplotypes have been associated with FM susceptibility and also pain sensitivity. Diatchenko et al.62 revealed that specific haplotypes of the COMT gene were associated with low, average, and high pain sensitivity, and the ACCG haplotype of the COMT gene was defined as a high pain sensitivity haplotype. Likewise, the ACCG haplotype of the COMT gene was more frequent among FM patients and was also associated with higher pain responsiveness to stimuli in a haplotype analysis.17 A previous report also showed associations between FM patients with the ACCG haplotype and higher FIQ scores.55
In conclusion, the COMT gene is one of most investigated genetic polymorphisms regarding susceptibility to FM. Although there are some negative results, COMT remains a highly likely candidate gene associated with FM susceptibility. Moreover, research is still in progress to evaluate the role of COMT gene polymorphisms in the disease course and the outcomes of FM. Further prospective largescale studies are needed to connect clinical practice and the development of drugs for FM.
Dopamine is a key neurotransmitter involved in modulating pain perception and natural analgesia.63 Furthermore, dopamine has an important role in the descending inhibition of pain.64 In fact, altered dopamine receptor function has been demonstrated in FM patients.6566 Malt et al.65 reported that FM patients showed an increased prolactin response (dopamine sensitivity) to the buspirone challenge test. But at the same time, FM patients presented no difference in hypothermic response or growth hormone release (5HT sensitivity), which suggests altered dopaminergic neurotransmission among FM patients. Moreover, pramipexole, a dopamine-D2/D3 agonist, demonstrated higher efficacy in FM patients compared with placebo. Treatment with pramipexole showed improvement on scores used to evaluate pain, function, fatigue, and global status in FM.67 It is in this context that researchers have investigated dopaminergic receptor genes as potential contributors to genetic susceptibility to FM. Buskila et al.26 reported that the D4 dopamine receptor exon III repeat polymorphism was associated with the personality profile of FM patients. FM patients with those polymorphisms scored low on the TPQ novelty-seeking personality trait.
Again, there remains insufficient research to fully understand the role of the dopaminergic receptor genes in FM. However, on the basis of the effects of pramipexole in FM, it is worth further evaluating dopaminergic-related gene polymorphisms in FM.
The genetic factors discussed so far do not fully explain the etiology of FM. Thus, researchers are also interested in other possible genetic polymorphisms. Because several ion channels contribute to the detection and processing of thermal, mechanical, and chemical pain stimuli,68 dysfunctional ion channels have been proposed as a possible risk factor associated with FM susceptibility. For example, one study suggested that the SCN9A gene-encoded dorsal root ganglia sodium channel polymorphism was associated with severe symptoms of FM.10
Recently, Kim et al.69 showed that certain GCH1 haplotypes had a protective role against FM susceptibility and pain sensitivity, suggesting that nitric oxide (NO) may be involved in pain sensitivity in the pathogenesis of FM. Oxidative stress and NO metabolism participate in various processes, such as vascular dilatation, neurotransmission, and immune regulation, and may also be involved in the regulation of pain in the pathogenesis of FM.7071
FM patients frequently have symptoms related to relentless sympathetic hyperactivity, which may also explain FM symptoms unrelated to pain sensitivity.72 Adrenergic receptors (ARs) act as binding sites for catecholamines and serve important roles in the responses of the sympathetic nervous system.73 Given this background, Vargas-Alarcón et al.74 investigated the association between AR gene polymorphism and FM and found that AR gene polymorphisms were associated with the risk of developing FM and were also related to different domains of the FIQ score.
The pathophysiology and symptoms of FM are not fully described by the genetic factors highlighted in this review, even if all of the potential contributions discussed here are confirmed through further research. Thus, to deepen our understanding of the genetic basis of FM, researchers have also examined other pain-related genes. GWAS or genetic studies performed in other FM-related disorders would also be of interest.
Go to : 
The pathogenesis of FM is not yet fully understood. However, FM is currently thought to result from the interaction of numerous factors including physical, psychological, genetic, neurobiological, and environmental factors. Because familial aggregation strongly supports the likelihood of a genetic component to the etiology of FM, recent research has focused on genetic susceptibility and environmental exposures that trigger further alterations in gene expression.
In this context, certain polymorphisms involving genes from the serotonergic, dopaminergic, and catecholaminergic systems have been suggested as possible risk factors for the development of FM. Additionally, these polymorphisms may influence symptom severity and could be targets for pharmacological treatments. However, these genetic factors do not fully explain the etiology of FM. Moreover, researchers have as yet paid little attention to the effects of genetic factors on disease progression and treatment outcomes of FM. Further studies are required to evaluate the effects of genetic polymorphism on clinical outcomes in patients with FM. Indeed, many more prospective studies, involving larger numbers of FM patients, are needed to expand our understanding of the role of genetics in FM.
Go to : 
ACKNOWLEDGEMENTS
This study was supported by a grant (CRI 14033-1) from Chonnam National University Hospital Biomedical Research Institute in 2014.
Go to : 
References
1. Jones GT, Atzeni F, Beasley M, Flüβ E, Sarzi-Puttini P, Macfarlane GJ. The prevalence of fibromyalgia in the general population: a comparison of the American College of Rheumatology 1990, 2010, and modified 2010 classification criteria. Arthritis Rheumatol. 2015; 67:568–575. PMID: 25323744.
2. Bennett RM, Friend R, Jones KD, Ward R, Han BK, Ross RL. The Revised Fibromyalgia Impact Questionnaire (FIQR): validation and psychometric properties. Arthritis Res Ther. 2009; 11:R120. PMID: 19664287.
3. Wolfe F, Clauw DJ, Fitzcharles MA, Goldenberg DL, Katz RS, Mease P, et al. The American College of Rheumatology preliminary diagnostic criteria for fibromyalgia and measurement of symptom severity. Arthritis Care Res (Hoboken). 2010; 62:600–610. PMID: 20461783.
4. Kim SK, Kim SH, Lee CK, Lee HS, Lee SH, Park YB, et al. Effect of fibromyalgia syndrome on the health-related quality of life and economic burden in Korea. Rheumatology (Oxford). 2013; 52:311–320. PMID: 23024016.
5. Doron Y, Peleg R, Peleg A, Neumann L, Buskila D. The clinical and economic burden of fibromyalgia compared with diabetes mellitus and hypertension among Bedouin women in the Negev. Fam Pract. 2004; 21:415–419. PMID: 15249530.
6. Maletic V, Raison CL. Neurobiology of depression, fibromyalgia and neuropathic pain. Front Biosci (Landmark Ed). 2009; 14:5291–5338. PMID: 19482616.
7. Sommer C, Häuser W, Burgmer M, Engelhardt R, Gerhold K, Petzke F, et al. Arbeitsgemeinschaft der Wissenschaftlichen Medizinischen Fachgesellschaften. Etiology and pathophysiology of fibromyalgia syndrome. Schmerz. 2012; 26:259–267. PMID: 22760458.
8. Ablin J, Neumann L, Buskila D. Pathogenesis of fibromyalgia -a review. Joint Bone Spine. 2008; 75:273–279. PMID: 18375167.
9. Buskila D, Sarzi-Puttini P. Biology and therapy of fibromyalgia. Genetic aspects of fibromyalgia syndrome. Arthritis Res Ther. 2006; 8:218. PMID: 16887010.
10. Vargas-Alarcon G, Alvarez-Leon E, Fragoso JM, Vargas A, Martinez A, Vallejo M, et al. A SCN9A gene-encoded dorsal root ganglia sodium channel polymorphism associated with severe fibromyalgia. BMC Musculoskelet Disord. 2012; 13:23. PMID: 22348792.
11. Mogil JS. Pain genetics: past, present and future. Trends Genet. 2012; 28:258–266. PMID: 22464640.
12. Altman D, Lundholm C, Milsom I, Peeker R, Fall M, Iliadou AN, et al. The genetic and environmental contribution to the occurrence of bladder pain syndrome: an empirical approach in a nationwide population sample. Eur Urol. 2011; 59:280–285. PMID: 21056533.
13. Peters MJ, Broer L, Willemen HL, Eiriksdottir G, Hocking LJ, Holliday KL, et al. Genome-wide association study meta-analysis of chronic widespread pain: evidence for involvement of the 5p15.2 region. Ann Rheum Dis. 2013; 72:427–436. PMID: 22956598.
14. Wang W, Gu J, Li YQ, Tao YX. Are voltage-gated sodium channels on the dorsal root ganglion involved in the development of neuropathic pain? Mol Pain. 2011; 7:16. PMID: 21345196.
15. Ichinose H, Suzuki T, Inagaki H, Ohye T, Nagatsu T. Molecular genetics of dopa-responsive dystonia. Biol Chem. 1999; 380:1355–1364. PMID: 10661862.
16. Oertel B, Lötsch J. Genetic mutations that prevent pain: implications for future pain medication. Pharmacogenomics. 2008; 9:179–194. PMID: 18370847.
17. Martínez-Jauand M, Sitges C, Rodríguez V, Picornell A, Ramon M, Buskila D, et al. Pain sensitivity in fibromyalgia is associated with catechol-O-methyltransferase (COMT) gene. Eur J Pain. 2013; 17:16–27. PMID: 22528689.
18. Pellegrino MJ, Waylonis GW, Sommer A. Familial occurrence of primary fibromyalgia. Arch Phys Med Rehabil. 1989; 70:61–63. PMID: 2916922.
19. Wolfe F, Smythe HA, Yunus MB, Bennett RM, Bombardier C, Goldenberg DL, et al. The American College of Rheumatology 1990 criteria for the classification of fibromyalgia. Report of the Multicenter Criteria Committee. Arthritis Rheum. 1990; 33:160–172. PMID: 2306288.
20. Buskila D, Neumann L, Hazanov I, Carmi R. Familial aggregation in the fibromyalgia syndrome. Semin Arthritis Rheum. 1996; 26:605–611. PMID: 8989805.
21. Buskila D, Neumann L. Fibromyalgia syndrome (FM) and nonarticular tenderness in relatives of patients with FM. J Rheumatol. 1997; 24:941–944. PMID: 9150086.
22. Arnold LM, Hudson JI, Hess EV, Ware AE, Fritz DA, Auchenbach MB, et al. Family study of fibromyalgia. Arthritis Rheum. 2004; 50:944–952. PMID: 15022338.
23. Fitzcharles MA, Rampakakis E, Ste-Marie PA, Sampalis JS, Shir Y. The association of socioeconomic status and symptom severity in persons with fibromyalgia. J Rheumatol. 2014; 41:1398–1404. PMID: 24931954.
24. Assumpção A, Cavalcante AB, Capela CE, Sauer JF, Chalot SD, Pereira CA, et al. Prevalence of fibromyalgia in a low socioeconomic status population. BMC Musculoskelet Disord. 2009; 10:64. PMID: 19505321.
25. Cohen H, Buskila D, Neumann L, Ebstein RP. Confirmation of an association between fibromyalgia and serotonin transporter promoter region (5-HTTLPR) polymorphism, and relationship to anxiety-related personality traits. Arthritis Rheum. 2002; 46:845–847. PMID: 11920428.
26. Buskila D, Cohen H, Neumann L, Ebstein RP. An association between fibromyalgia and the dopamine D4 receptor exon III repeat polymorphism and relationship to novelty seeking personality traits. Mol Psychiatry. 2004; 9:730–731. PMID: 15052273.
27. Gürsoy S, Erdal E, Herken H, Madenci E, Alaşehirli B. Association of T102C polymorphism of the 5-HT2A receptor gene with psychiatric status in fibromyalgia syndrome. Rheumatol Int. 2001; 21:58–61. PMID: 11732859.
28. Finan PH, Zautra AJ, Davis MC, Lemery-Chalfant K, Covault J, Tennen H. COMT moderates the relation of daily maladaptive coping and pain in fibromyalgia. Pain. 2011; 152:300–307. PMID: 21130573.
29. Norrgard K. Genetic variation and disease: GWAS. Nat Educ. 2008; 1:87.
30. Landstrom AP, Ackerman MJ. GWAS or Gee Whiz, PSAS or Pshaw: elucidating the biologic and clinical significance of genetic variation in cardiovascular disease. Heart Rhythm. 2009; 6:1751–1753. PMID: 19959124.
31. Dawn Teare M, Barrett JH. Genetic linkage studies. Lancet. 2005; 366:1036–1044. PMID: 16168786.
32. Docampo E, Escaramís G, Gratacòs M, Villatoro S, Puig A, Kogevinas M, et al. Genome-wide analysis of single nucleotide polymorphisms and copy number variants in fibromyalgia suggest a role for the central nervous system. Pain. 2014; 155:1102–1109. PMID: 24582949.
33. Feng J, Zhang Z, Wu X, Mao A, Chang F, Deng X, et al. Discovery of potential new gene variants and inflammatory cytokine associations with fibromyalgia syndrome by whole exome sequencing. PLoS One. 2013; 8:e65033. PMID: 23762283.
34. Arnold LM, Fan J, Russell IJ, Yunus MB, Khan MA, Kushner I, et al. The fibromyalgia family study: a genome-wide linkage scan study. Arthritis Rheum. 2013; 65:1122–1128. PMID: 23280346.
35. Russell IJ, Michalek JE, Vipraio GA, Fletcher EM, Javors MA, Bowden CA. Platelet 3H-imipramine uptake receptor density and serum serotonin levels in patients with fibromyalgia/fibrositis syndrome. J Rheumatol. 1992; 19:104–109. PMID: 1313504.
36. Russell IJ, Vaeroy H, Javors M, Nyberg F. Cerebrospinal fluid biogenic amine metabolites in fibromyalgia/fibrositis syndrome and rheumatoid arthritis. Arthritis Rheum. 1992; 35:550–556. PMID: 1374252.
37. Wolfe F, Russell IJ, Vipraio G, Ross K, Anderson J. Serotonin levels, pain threshold, and fibromyalgia symptoms in the general population. J Rheumatol. 1997; 24:555–559. PMID: 9058665.
38. Kuzelova H, Ptacek R, Macek M. The serotonin transporter gene (5-HTT) variant and psychiatric disorders: review of current literature. Neuro Endocrinol Lett. 2010; 31:4–10. PMID: 20150867.
39. Bondy B, Spaeth M, Offenbaecher M, Glatzeder K, Stratz T, Schwarz M, et al. The T102C polymorphism of the 5-HT2A-receptor gene in fibromyalgia. Neurobiol Dis. 1999; 6:433–439. PMID: 10527809.
40. Lee YH, Choi SJ, Ji JD, Song GG. Candidate gene studies of fibromyalgia: a systematic review and meta-analysis. Rheumatol Int. 2012; 32:417–426. PMID: 21120487.
41. Offenbaecher M, Bondy B, de Jonge S, Glatzeder K, Krüger M, Schoeps P, et al. Possible association of fibromyalgia with a polymorphism in the serotonin transporter gene regulatory region. Arthritis Rheum. 1999; 42:2482–2488. PMID: 10555044.
42. Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996; 274:1527–1531. PMID: 8929413.
43. Gursoy S. Absence of association of the serotonin transporter gene polymorphism with the mentally healthy subset of fibromyalgia patients. Clin Rheumatol. 2002; 21:194–197. PMID: 12111622.
44. Frank B, Niesler B, Bondy B, Späth M, Pongratz DE, Ackenheil M, et al. Mutational analysis of serotonin receptor genes: HTR3A and HTR3B in fibromyalgia patients. Clin Rheumatol. 2004; 23:338–344. PMID: 15293096.
45. Hrycaj P, Stratz T, Mennet P, Müller W. Pathogenetic aspects of responsiveness to ondansetron (5-hydroxytryptamine type 3 receptor antagonist) in patients with primary fibromyalgia syndrome--a preliminary study. J Rheumatol. 1996; 23:1418–1423. PMID: 8856622.
46. Haus U, Varga B, Stratz T, Späth M, Müller W. Oral treatment of fibromyalgia with tropisetron given over 28 days: influence on functional and vegetative symptoms, psychometric parameters and pain. Scand J Rheumatol Suppl. 2000; 113:55–58. PMID: 11028833.
47. Ernberg M, Lundeberg T, Kopp S. Effects on muscle pain by intramuscular injection of granisetron in patients with fibromyalgia. Pain. 2003; 101:275–282. PMID: 12583870.
48. Murphy DL, Andrews AM, Wichems CH, Li Q, Tohda M, Greenberg B. Brain serotonin neurotransmission: an overview and update with an emphasis on serotonin subsystem heterogeneity, multiple receptors, interactions with other neurotransmitter systems, and consequent implications for understanding the actions of serotonergic drugs. J Clin Psychiatry. 1998; 59(Suppl 15):4–12. PMID: 9786305.
49. Latremoliere A, Woolf CJ. Central sensitization: a generator of pain hypersensitivity by central neural plasticity. J Pain. 2009; 10:895–926. PMID: 19712899.
50. Woolf CJ. Central sensitization: implications for the diagnosis and treatment of pain. Pain. 2011; 152(3 Suppl):S2–S15. PMID: 20961685.
51. Zubieta JK, Heitzeg MM, Smith YR, Bueller JA, Xu K, Xu Y, et al. COMT val158met genotype affects mu-opioid neurotransmitter responses to a pain stressor. Science. 2003; 299:1240–1243. PMID: 12595695.
52. Männistö PT, Kaakkola S. Catechol-O-methyltransferase (COMT): biochemistry, molecular biology, pharmacology, and clinical efficacy of the new selective COMT inhibitors. Pharmacol Rev. 1999; 51:593–628. PMID: 10581325.
53. Matsuda JB, Barbosa FR, Morel LJ, França Sde C, Zingaretti SM, da Silva LM, et al. Serotonin receptor (5-HT 2A) and catechol-O-methyltransferase (COMT) gene polymorphisms: triggers of fibromyalgia? Rev Bras Reumatol. 2010; 50:141–149. PMID: 21125150.
54. Gürsoy S, Erdal E, Herken H, Madenci E, Alaşehirli B, Erdal N. Significance of catechol-O-methyltransferase gene polymorphism in fibromyalgia syndrome. Rheumatol Int. 2003; 23:104–107. PMID: 12739038.
55. Vargas-Alarcón G, Fragoso JM, Cruz-Robles D, Vargas A, Vargas A, Lao-Villadóniga JI, et al. Catechol-O-methyltransferase gene haplotypes in Mexican and Spanish patients with fibromyalgia. Arthritis Res Ther. 2007; 9:R110. PMID: 17961261.
56. Cohen H, Neumann L, Glazer Y, Ebstein RP, Buskila D. The relationship between a common catechol-O-methyltransferase (COMT) polymorphism val(158) met and fibromyalgia. Clin Exp Rheumatol. 2009; 27(5 Suppl 56):S51–S56. PMID: 20074440.
57. Barbosa FR, Matsuda JB, Mazucato M, de Castro França S, Zingaretti SM, da Silva LM, et al. Influence of catechol-O-methyltransferase (COMT) gene polymorphisms in pain sensibility of Brazilian fibromialgia patients. Rheumatol Int. 2012; 32:427–430. PMID: 21120493.
58. Potvin S, Larouche A, Normand E, de Souza JB, Gaumond I, Grignon S, et al. DRD3 Ser9Gly polymorphism is related to thermal pain perception and modulation in chronic widespread pain patients and healthy controls. J Pain. 2009; 10:969–975. PMID: 19464960.
59. Tander B, Gunes S, Boke O, Alayli G, Kara N, Bagci H, et al. Polymorphisms of the serotonin-2A receptor and catechol-O-methyltransferase genes: a study on fibromyalgia susceptibility. Rheumatol Int. 2008; 28:685–691. PMID: 18196244.
60. Lee YH, Kim JH, Song GG. Association between the COMT Val158Met polymorphism and fibromyalgia susceptibility and fibromyalgia impact questionnaire score: a meta-analysis. Rheumatol Int. 2015; 35:159–166. PMID: 24951880.
61. International HapMap Consortium. A haplotype map of the human genome. Nature. 2005; 437:1299–1320. PMID: 16255080.
62. Diatchenko L, Nackley AG, Slade GD, Bhalang K, Belfer I, Max MB, et al. Catechol-O-methyltransferase gene polymorphisms are associated with multiple pain-evoking stimuli. Pain. 2006; 125:216–224. PMID: 16837133.
63. Jarcho JM, Mayer EA, Jiang ZK, Feier NA, London ED. Pain, affective symptoms, and cognitive deficits in patients with cerebral dopamine dysfunction. Pain. 2012; 153:744–754. PMID: 22386471.
64. Wood PB. Role of central dopamine in pain and analgesia. Expert Rev Neurother. 2008; 8:781–797. PMID: 18457535.
65. Malt EA, Olafsson S, Aakvaag A, Lund A, Ursin H. Altered dopamine D2 receptor function in fibromyalgia patients: a neuroendocrine study with buspirone in women with fibromyalgia compared to female population based controls. J Affect Disord. 2003; 75:77–82. PMID: 12781354.
66. Wood PB, Schweinhardt P, Jaeger E, Dagher A, Hakyemez H, Rabiner EA, et al. Fibromyalgia patients show an abnormal dopamine response to pain. Eur J Neurosci. 2007; 25:3576–3582. PMID: 17610577.
67. Holman AJ, Myers RR. A randomized, double-blind, placebo-controlled trial of pramipexole, a dopamine agonist, in patients with fibromyalgia receiving concomitant medications. Arthritis Rheum. 2005; 52:2495–2505. PMID: 16052595.
68. Eglen RM, Hunter JC, Dray A. Ions in the fire: recent ion-channel research and approaches to pain therapy. Trends Pharmacol Sci. 1999; 20:337–342. PMID: 10431213.
69. Kim SK, Kim SH, Nah SS, Lee JH, Hong SJ, Kim HS, et al. Association of guanosine triphosphate cyclohydrolase 1 gene polymorphisms with fibromyalgia syndrome in a Korean population. J Rheumatol. 2013; 40:316–322. PMID: 23322459.
70. Kingwell BA. Nitric oxide-mediated metabolic regulation during exercise: effects of training in health and cardiovascular disease. FASEB J. 2000; 14:1685–1696. PMID: 10973917.
71. Larson AA, Giovengo SL, Russell IJ, Michalek JE. Changes in the concentrations of amino acids in the cerebrospinal fluid that correlate with pain in patients with fibromyalgia: implications for nitric oxide pathways. Pain. 2000; 87:201–211. PMID: 10924813.
72. Martinez-Lavin M. Biology and therapy of fibromyalgia. Stress, the stress response system, and fibromyalgia. Arthritis Res Ther. 2007; 9:216. PMID: 17626613.
73. Small KM, McGraw DW, Liggett SB. Pharmacology and physiology of human adrenergic receptor polymorphisms. Annu Rev Pharmacol Toxicol. 2003; 43:381–411. PMID: 12540746.
74. Vargas-Alarcón G, Fragoso JM, Cruz-Robles D, Vargas A, Martinez A, Lao-Villadóniga JI, et al. Association of adrenergic receptor gene polymorphisms with different fibromyalgia syndrome domains. Arthritis Rheum. 2009; 60:2169–2173. PMID: 19565482.
Go to : 




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download