Abstract
Dysregulation of microRNA (miRNA) levels is observed in diverse disease states. Early studies showed that by analyzing the expression profile of miRNAs in the tissue sample of a diseased person, it was possible to classify the disease into a specific subtype. To be used for diagnostic purposes more practically, however, a less invasive method than tissue biopsy is required. Surprisingly, it was discovered that a notable amount of extracellular miRNAs circulate throughout the body fluids with high stability. Moreover, the expression profile of miRNAs was shown to differ considerably between healthy and diseased people. In addition, evidence has been accumulating of extracellular miRNAs acting as signaling molecules between distantly located cells. If the expression profile faithfully reflects the disease states, the profiling of extracellular miRNAs will become a useful means of early warning or diagnosis of diverse diseases, replacing more invasive biopsy methods.
MicroRNAs (miRNAs) are small (~22 nucleotides) noncoding RNAs that regulate more than half of the genes in human cells.1 By complementary binding into the 3'-untranslated region (3'-UTR) of mRNAs, they suppress translation and induce degradation of mRNAs. Because of their broad targeting range, miRNAs are expected to be involved in most cellular processes. Accordingly, dysregulation of the expression of miRNAs results in diverse disease states.2
Analysis of miRNA levels in human plasma and serum showed that extracellular miRNAs are unexpectedly stable in body fluids.3 Moreover, many studies showed that the miRNA profile is altered in the blood of patients compared with that of healthy persons. These discoveries have attracted much interest in utilizing the level of extracellular miRNAs as a novel biomarker in diverse diseases.
In this review, I summarize recent studies about extracellular miRNAs that have been analyzed as disease markers. I briefly introduce in chronological order the important discoveries that led to the establishment of the miRNAs as novel biomarkers. I also discuss the possible origin of extracellular miRNAs and the reliability of miRNAs as biomarkers. Finally, I make several suggestions for experimental processes to identify miRNA biomarkers with higher consistency. Because of the many such studies, reports of specific miRNAs for disease markers are not dealt with thoroughly in this review. I recommend that researchers who want to identify novel biomarkers or who want to search the identified markers for a specific disease search the databases in which the published miRNA markers for diverse diseases are collected, with links to their corresponding published studies.4 Note that although diverse terms are used to define miRNA in the extracellular space, including extracellular miRNA, vesicular miRNA, exosomal miRNA, and circulating miRNA, I will use the term extracellular miRNA to represent all miRNAs detected outside cells.
After the discovery of the first miRNA in Caenorhabditis elegans in 1993, it took 7 years to encounter the second miRNA, also in C. elegans (Fig. 1).567 Until that time, these tiny RNAs were regarded as molecules specific to lower organisms. The importance of miRNA as a regulatory molecule was first implied by the discovery that the second miRNA identified, let-7, is conserved throughout bilateral animals.8 The identification of dozens of miRNAs from worm, fly, and human suggested that miRNAs indeed constitute a large group of previously unknown regulatory molecules.91011 The function of miRNAs began to be discovered, and it was soon reported that miRNAs are highly related to human diseases, such as chronic myeloid leukemia (CLL).12 Numerous studies to analyze the involvement of miRNAs in human disease were triggered by the development of the microarray platform, which made the global profiling of miRNA possible.13 Strikingly, the classification of cancer phenotypes was more successful when the expression profile of miRNA rather than that of mRNA was used.13 The availability of profiling based on microarray, and subsequently next generation sequencing, established a foundation for those studies to discover miRNA biomarkers for the classification and diagnosis of disease (Fig. 1).
Although the analysis of the miRNA profile in human tissues showed the great potential of miRNA as a disease marker, a less invasive method would make this analysis more practical. In this regard, the identification of miRNAs in body fluids drew much attention. Extracellular miRNAs were first discovered in cell culture medium.14 Interestingly, encapsulated inside exosomes, the extracellular miRNAs secreted from donor cells could be transferred into other recipient cells. The existence of extracellular miRNAs in the body fluids of humans was soon reported in several studies.15161718 An important discovery was that the miRNA profiles in the body fluids of diseased persons were considerably different from the profiles in healthy persons.Moreover, the miRNAs in body fluids remained stable in diverse severe conditions such as boiling, very low or high pH, repetitive freezing-thawing, and storage at room temperature for a long time.1518 These studies raised the possibility that miRNAs from body fluids could be used to diagnose human diseases. After these discoveries, hundreds of research studies were conducted to find extracellular miRNAs that could be used as diagnostic or prognostic markers for specific diseases (Fig. 2). It is much easier to profile the global level of miRNAs rather than that of proteins or metabolites because of the high sensitivity and throughput of transcriptome profiling techniques. Furthermore, thousands of miRNAs with tissue- and disease-specific expression patterns exist in humans. For these reasons, a large amount of information can be obtained within a relatively short time when miRNAs are used as biomarkers.
In addition to these kinds of research, recent studies suggested that extracellular miRNAs may be used as active signaling molecules. It was reported that some miRNAs released from donor cells exert regulatory effects on the mRNAs in recipient cells, functioning as endocrine signals.19 Unexpectedly, both precursor miRNAs (pre-miRNAs) and the enzymes required for miRNA biogenesis were observed in the extracellular exosomes, enabling the production of miRNAs inside the exosomes.20 Moreover, those exosomes derived from the cells and sera of breast cancer patients could make normal cells become cancerous.20 These studies and many other reports support the hypothesis that miRNA can function as a previously unidentified hormone transferring signals between distant cells.21
One simple explanation for the origin of extracellular miRNAs is that the death of cells as a consequence of disease or other necrotic events results in the passive release of miRNAs in the cytoplasm, and that the released miRNAs are detected in the blood. Although this kind of miRNA release is feasible, it is likely that a regulatory pathway is also involved in the cellular release of miRNAs.
Initial study found that the miRNA profile inside cells differs from the miRNA profile in the culture media, which suggests the existence of a selective pathway for the release of miRNAs.14 miRNAs are found in multivesicular bodies (MVBs), the cellular structure containing vesicles that are released into the extracellular space as exosomes (Fig. 3A).22 Later, it was shown that neutral sphingomyelinase 2 (nSMase2) is involved in the secretion of miRNA-containing exosomes into the extracellular space.23 The secretion of exosomes is triggered by ceramides, whose synthesis is regulated by nSMase2. Also, heterogeneous nuclear ribonucleoprotein A2B1 (hnRNPA2B1) was identified to recognize specific sequence motifs in a subset of miRNAs and to regulate their sorting into exosomes.24 Although these studies identified the factors involved in the release of cellular miRNAs into the extracellular space, we still do not understand the detailed mechanism governing this process. The elucidation of the mechanism by which miRNAs are selected to be released from donor cells, how those specific groups of miRNAs are embedded inside the exosomes, and when the release of miRNAs is triggered would be helpful for studies attempting to use extracellular miRNAs as biomarkers in diseases.
Extracellular miRNAs have been discovered in diverse macromolecules in the blood and other body fluids (Fig. 3B). Because of their small size, miRNAs can be incorporated into some proteins and protected from nuclease attack. Ago protein is the most prevalent such protein, because normal biogenesis results in the incorporation of miRNA into the Ago protein. The majority of extracellular miRNAs cofractionate with Ago2 protein complexes, whereas only a small portion of miRNAs are encapsulated in the extracellular vesicles or exosomes in the human plasma.2526 In the bloodstream, various extracellular vesicles or exosomes circulate throughout the body. The extracellular vesicles are made by direct budding of the cellular membrane, whereas the exosomes are released from the cells as a result of the fusion of MVBs into the plasma membrane.27 Early studies identified diverse miRNAs in these extracellular structures (Fig. 3B).2829 In addition, many following studies determined the level of those miRNAs and showed that the miRNA level could be used to predict whether a person has a disease or not.3
Another interesting discovery was that extracellular miRNAs are embedded in high-density lipoprotein (HDL).30 The expression profile of miRNAs in HDL showed that the miRNA population clearly differs between samples from healthy persons and samples from persons with atherosclerosis. Moreover, the nSMase2-dependent pathway was noted to be involved in the release of miRNA-containing HDL from the cells.30 More studies are required to determine whether the alteration of the miRNA level in HDL is linked to disease progression.
In a recent study, it was shown that vesicle-embedded miRNAs injected into the mouse bloodstream could be distributed into diverse tissues and suppress target mRNAs in recipient cells.31 Moreover, direct in vivo evidence of RNA exchange between cells through exosomes was presented in living mice.32 These studies confirmed that miRNAs actually circulate throughout the body fluids and function in targeted cells.
Compared with the mechanism of the release of miRNA-containing macromolecules, there are no reports regarding the mechanism of uptake. Although the receptor for the miRNA-containing exosomes or miRNA-Ago2 complexes has not been identified, it is possible that the specific uptake by target cells might be determined by the presence of a cell surface receptor for those macromolecules. It is also plausible that the specificity is determined by the existence of target mRNAs in the recipient cells, because only the mRNAs containing miRNA-binding sites would be influenced. More research is required to elucidate the mechanism of both cellular release and uptake of miRNAs.
Although most studies to identify miRNA biomarkers use blood, plasma, or serum, miRNAs are observed in most body fluids (Fig. 3C).433 Because different miRNAs are profiled from each body fluid, researchers should select the proper fluid sample for their purpose. Widespread distribution of miRNAs in the body fluids suggests that miRNA biomarkers could be applicable for diverse diseases.
Compared to currently used biomarkers, which are generally based on the levels of specific proteins in the blood, a novel biomarker based on miRNA levels has several merits. One of the strongest points is that the global miRNA level can be measured rapidly and accurately. Current sequencing technology enables the analysis of billions of sequences in a single run with very high precision.34 The entire miRNA population, which is composed of thousands of different sequences in humans, can be rapidly measured owing to high-throughput sequencing technology.35 Because diverse miRNAs are expressed differentially in different tissues and cellular states, the combination of miRNA levels would provide a wealth of information. An early study designed to classify the origin of cancer tissues on the basis of the miRNA level showed that the combination of only 48 miRNAs was enough to reach near perfect accuracy in the classification of most cancer tissues.36 Although a similar study has not been systematically performed using miRNA levels in body fluids, it is worth trying a similar approach with the combination of thousands of miRNAs in the body fluids.
For a new biomarker to be utilized, it is essential to have a reagent or a method that can be used to measure the level of the marker with high specificity. Compared with the technique used to detect nucleic acids, it is more laborious and time-consuming to develop a specific antibody or other kind of detection agent against a protein-based marker. On the other hand, with the help of the robust polymerase chain reaction (PCR) and sequencing technologies, there is nearly no need to develop a new agent for the detection of novel RNA molecules.
Although various biomolecules containing useful information for a disease state may exist in a sample, they would be less reliable if the amount of starting material is too small and the quantity of the molecules cannot be measured. In this sense, nucleic acid-based measurement has strong merit because of the availability of amplification. Compared with the measurement of protein or other kinds of molecules, for which no amplification method is applicable, PCR-based amplification enables the measurement of very low amounts of miRNAs in a small volume of body fluids.
Finally, the high stability of miRNAs in the body fluids makes miRNA a good candidate for a biomarker. As discussed above, miRNAs in the blood are protected from nuclease-mediated cleavage. Other typical longer RNAs such as messenger RNAs are too long to be protected inside protein complexes or small vesicles. In summary, the diversity and stability of miRNAs in combination with technical availabilities establish miRNAs as novel and promising biomarkers for the diagnosis of human diseases.
Although many new miRNAs that can be used as disease markers have been identified in body fluids, more studies are needed before miRNAs can be used as reliable biomarkers. One of the most critical problems in utilizing miRNAs as disease markers is the lack of consistency between reports. That is, different and nonoverlapping sets of miRNAs have been reported as biomarkers for the same disease. In a study that collected data from several reports in which the miRNA level was measured in plasma and serum to identify breast cancer-specific markers, there were no overlapping miRNAs among all the previously published papers.37 Although the exact reason for this discrepancy was not scrutinized, a major reason may be methodological differences in the experimental processes.38 To enable the discovery of more useful miRNA biomarkers of diseases and to increase their consistency, several methodological aspects should be considered.
First, although body fluids contain diverse macromolecules of different composition, subpopulation-specific RNA preparation is not considered carefully. In a previous study, for example, it was shown that miRNA profiles differ considerably between serum and plasma.39 The suspected reason for this difference was that the miRNAs were released from blood cells into serum during coagulation. Also, different sets of miRNAs are observed in different populations of extracellular vesicles and exosomes. For example, it was shown that different molecules were profiled between exosomes that were morphologically similar but that differed in surface antigens.40 Extracellular vesicles of different sizes also have different contents. For example, one kind of exosome called an oncosome is larger than typical exosomes and is similar in size to platelets.41 It was reported that a specific group of miRNAs enriched in the oncosome enhanced the cancer phenotype. In addition, miRNAs are also embedded inside different protein complexes. Because of these diversities, the miRNA profile can be influenced by the choice of subpopulation in the body fluids.
Second, because the amount of miRNAs inside the body fluids is small, the profiling of miRNAs is greatly affected by minute perturbations of experimental processes. For example, we showed that miRNA profiles differed considerably when the initial amounts of total RNA differed, even though the same RNA was used for miRNA extraction.42 This is because miRNAs are small in size and their secondary structures are strikingly different, resulting in different degrees of precipitation during RNA extraction. Because it is not easy to standardize the initial amounts of RNA among fluid samples, miRNAs could be extracted with different efficiency from different body fluid samples.
Third, there is still no proper control for the normalization of the miRNA level in the body fluids. The control RNAs generally used for measurement of the cellular miRNA level, which include U6 small nuclear RNA (snRNA) and 5S ribosomal RNA (rRNA), do not give consistent results when used for extracellular miRNA control.15 Moreover, there is no known miRNA with a consistent expression level among the diverse body fluids. This problem makes the quantitation of the miRNA level and the comparison of miRNA levels between published data difficult.
To overcome the aspects described above, diverse factors should be considered during the preparation of samples and the analysis of results. Standardized sampling and processing protocols are required, including sample preparation methods, RNA extraction methods, and profiling platforms. In addition, one should make comparisons only between published data derived from experiments conducted with the use of the same protocol. Also, detailed protocols for complete processes should be described in the text for later comparison by other researchers.
Because a small amount of cell debris contains a large quantity of RNA molecules, it is necessary to ensure that the samples are not contaminated by blood cells during RNA extraction process.43 Excessive mechanical force should be avoided during the removal of cells from the blood. Even after excluding the possibility of blood cell contamination, one should be cautious when the identified biomarker is one of the well-known blood cell-enriched miRNAs. The suspicious miRNAs originating from blood cells include miR-150 from lymphoid blood cells; let-7a, miR-197, miR-223, and miR-574-3p from myeloid blood cells; and miR-16, miR-92a, miR-451, and miR-486-5p from red blood cells.43 It is better to exclude these miRNAs.
During RNA preparation from blood samples, diverse chemicals are used to prevent coagulation. These anticoagulants include EDTA, heparin, and citrate. Differences in the use of anticoagulants may result in different miRNA profiles. Of the anticoagulants, EDTA has a minimal effect on the miRNA profile during the sample preparation step.38 On the other hand, heparin and citrate inhibit the PCR, which is the inevitable step for amplifying the small amount of miRNAs in body fluid samples.
Because the plasma or serum contains diverse sets of macromolecules, results will be more reliable if researchers measure the expression level of miRNAs in a specific subpopulation. Possible choices of macromolecules include exosomes, extracellular vesicles, HDL, and Ago protein. The main drawback in purifying a subpopulation is that the experimental process is somewhat laborious and time-consuming. To circumvent this problem, diverse commercial fractionation kits used to enrich a specific subpopulation in the blood are available. However, controversy exists over whether the data produced from those kits are more reliable than the data obtained by the traditional ultracentrifuge-based method.44 More studies and technical advances are required to make the purification of a subpopulation easier. It would also be better if one could purify the extracellular vesicles or exosomes originating from a specific tissue, namely, a diseased tissue. It is plausible that proteins or lipids in the membrane of the vesicle may vary among those vesicles originating from different types of cells, although more studies are required to determine whether this kind of approach is feasible.
Caution is also needed when the discovered biomarker is known to be expressed ubiquitously. Some miRNAs were repeatedly observed to be increased or decreased in the blood of patients with different types of diseases. For example, the level of extracellular miR-21 is known to increase in many diseases, including diverse cancer types, hepatitis, multiple sclerosis, myocardial infarction, and lupus.45 Other representative miRNAs with expression changes in diverse diseases include miR-16, miR-126, miR-146a, miR-155, and miR-223. In the case of miRNAs included in this list, the expression change of a single miRNA would not be enough for disease classification. Instead, the combination of expression changes based on multiple miRNAs would be more reliable as a biomarker.
Because of the lack of proper endogenous control for normalization, it is recommended to add spike-in RNAs, the RNA molecules used to normalize measurements, into the same amount of body fluid samples right after the RNasedenaturation step. There is no known endogenous RNA type with a similar size of miRNAs. Several RNAs used for normalization such as U6 snRNA or 5S rRNA are much more susceptible to degradation than are miRNAs. Representative spike-in RNA is the synthetic miRNA with the sequence from organisms other than human. Examples of such spike-in RNAs, which are commercially available, include miR-39, miR-67, and miR-239b in C. elegans.
The body fluids may contain not only the miRNAs that are altered by disease states, but also other miRNAs that are secreted from diverse cells for the purpose of endocrine signals. As a result, it might be difficult to distinguish the disease-specific miRNA biomarkers from other molecules. In this case, the identification of miRNAs that are not detected in the body fluids under normal conditions, but are detected only in a specific disease state, could be a good strategy.
Since the discovery of miRNAs in body fluids, many efforts have been made to find reliable biomarkers for diverse diseases on the basis of the miRNA level. Despite many new discoveries about extracellular miRNAs, however, more research is needed to increase the reliability of miRNAs as promising disease markers. Because the experimental method for measuring the amount of miRNAs in the body fluids is still not optimized, more technical advances are needed. Moreover, many gaps still remain in our understanding about the physiological role of extracellular miRNAs and the mechanism of cellular release and uptake of miRNA-containing molecules. With the complementation of experimental techniques and the increase in our knowledge about extracellular miRNAs, the use of miRNAs as noninvasive biomarkers will become an invaluable tool.
Figures and Tables
 | FIG. 1Timeline of the important discoveries about the use of miRNAs as biomarkers. See text for details. |
 | FIG. 2Numbers of papers published each year about miRNA, miRNA biomarkers, and extracellular miRNA biomarkers, respectively. Only research papers were counted in PubMed. To count the papers about miRNA (solid line), papers containing the term "microRNA" or "microRNAs" in the title or abstract were searched. For papers about miRNA as a biomarker (gray bar), papers containing the term "biomarker" or "biomarkers" in the title or abstract were counted among the miRNA papers. For the papers about miRNA as an extracellular biomarker (black bar), papers were counted that additionally contained the term "blood-based", "extracellular", "exosome", "exosomes", "plasma", or "serum" among the papers about miRNA biomarkers. |
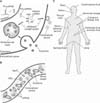 | FIG. 3Origin and distribution of extracellular miRNAs. (A) Release of miRNAs into the extracellular space. After the production of mature miRNAs, some portion of miRNAs might be incorporated into the multivesicular body and released into the extracellular space through exosomes. Another portion of miRNAs might be released directly through budding vesicles. (B) Circulating miRNAs in the blood vessels. The miRNAs, incorporated in the proteins such as Ago2, could circulate through the blood. It was also found that miRNAs exist in the extracellular vesicle, the exosome, and high-density lipoprotein (HDL). (C) miRNA-containing body fluids. Most of the body fluids were found to contain miRNAs. |
ACKNOWLEDGEMENTS
This work was supported by a research grant from the Research Institute of Medical Sciences, Chonnam National University (2015-CURIMS-DR002).
References
3. Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014; 11:145–156.

4. Russo F, Di Bella S, Nigita G, Macca V, Laganà A, Giugno R, et al. miRandola: extracellular circulating microRNAs database. PLoS One. 2012; 7:e47786.

5. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993; 75:843–854.

6. Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993; 75:855–862.

7. Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000; 403:901–906.

8. Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000; 408:86–89.

9. Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001; 294:853–858.

10. Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001; 294:858–862.

11. Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001; 294:862–864.

12. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002; 99:15524–15529.

13. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005; 435:834–838.

14. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007; 9:654–659.

15. Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008; 18:997–1006.

16. Chim SS, Shing TK, Hung EC, Leung TY, Lau TK, Chiu RW, et al. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008; 54:482–490.

17. Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008; 141:672–675.

18. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008; 105:10513–10518.

19. Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010; 39:133–144.

20. Melo SA, Sugimoto H, O'Connell JT, Kato N, Villanueva A, Vidal A, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014; 26:707–721.

21. Rottiers V, Näär AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012; 13:239–250.

22. Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009; 11:1143–1149.

23. Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010; 285:17442–17452.

24. Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Heránndez D, Vázquez J, Martin-Cofreces N, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013; 4:2980.

25. Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011; 108:5003–5008.

26. Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011; 39:7223–7233.

27. Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014; 30:255–289.

28. Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008; 3:e3694.

29. Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008; 110:13–21.

30. Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011; 13:423–433.

31. Bala S, Csak T, Momen-Heravi F, Lippai D, Kodys K, Catalano D, et al. Biodistribution and function of extracellular miRNA-155 in mice. Sci Rep. 2015; 5:10721.

32. Zomer A, Maynard C, Verweij FJ, Kamermans A, Schäfer R, Beerling E, et al. In Vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell. 2015; 161:1046–1057.

33. Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010; 56:1733–1741.

34. Reuter JA, Spacek DV, Snyder MP. High-throughput sequencing technologies. Mol Cell. 2015; 58:586–597.

35. Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014; 42(Database issue):D68–D73.

36. Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008; 26:462–469.

37. Witwer KW. Circulating microRNA biomarker studies: pitfalls and potential solutions. Clin Chem. 2015; 61:56–63.

38. Moldovan L, Batte KE, Trgovcich J, Wisler J, Marsh CB, Piper M. Methodological challenges in utilizing miRNAs as circulating biomarkers. J Cell Mol Med. 2014; 18:371–390.

39. Wang K, Yuan Y, Cho JH, McClarty S, Baxter D, Galas DJ. Comparing the MicroRNA spectrum between serum and plasma. PLoS One. 2012; 7:e41561.

40. Tauro BJ, Greening DW, Mathias RA, Mathivanan S, Ji H, Simpson RJ. Two distinct populations of exosomes are released from LIM1863 colon carcinoma cell-derived organoids. Mol Cell Proteomics. 2013; 12:587–598.

41. Morello M, Minciacchi VR, de Candia P, Yang J, Posadas E, Kim H, et al. Large oncosomes mediate intercellular transfer of functional microRNA. Cell Cycle. 2013; 12:3526–3536.

42. Kim YK, Yeo J, Kim B, Ha M, Kim VN. Short structured RNAs with low GC content are selectively lost during extraction from a small number of cells. Mol Cell. 2012; 46:893–895.

43. Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila). 2012; 5:492–497.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download