Abstract
After renal injury, selective damage occurs in the proximal tubules as a result of inhibition of glycolysis. The molecular mechanism of damage is not known. Poly(ADP-ribose) polymerase (PARP) activation plays a critical role of proximal tubular cell death in several renal disorders. Here, we studied the role of PARP on glycolytic flux in pig kidney proximal tubule epithelial LLC-PK1 cells using XFp extracellular flux analysis. Poly(ADP-ribosyl)ation by PARP activation was increased approximately 2-fold by incubation of the cells in 10 mM glucose for 30 minutes, but treatment with the PARP inhibitor 3-aminobenzamide (3-AB) does-dependently prevented the glucose-induced PARP activation (approximately 14.4% decrease in 0.1 mM 3-AB–treated group and 36.7% decrease in 1 mM 3-AB–treated group). Treatment with 1 mM 3-AB significantly enhanced the glucose-mediated increase in the extracellular acidification rate (61.1±4.3 mpH/min vs. 126.8±6.2 mpH/min or approximately 2-fold) compared with treatment with vehicle, indicating that PARP inhibition increases only glycolytic activity during glycolytic flux including basal glycolysis, glycolytic activity, and glycolytic capacity in kidney proximal tubule epithelial cells. Glucose increased the activities of glycolytic enzymes including hexokinase, phosphoglucose isomerase, phosphofructokinase-1, glyceraldehyde-3-phosphate dehydrogenase, enolase, and pyruvate kinase in LLC-PK1 cells. Furthermore, PARP inhibition selectively augmented the activities of hexokinase (approximately 1.4-fold over vehicle group), phosphofructokinase-1 (approximately 1.6-fold over vehicle group), and glyceraldehyde-3-phosphate dehydrogenase (approximately 2.2-fold over vehicle group). In conclusion, these data suggest that PARP activation may regulate glycolytic activity via poly(ADP-ribosyl)ation of hexokinase, phosphofructokinase-1, and glyceraldehyde-3-phosphate dehydrogenase in kidney proximal tubule epithelial cells.
Poly(ADP-ribose) polymerase (PARP) is a nuclear protein that regulates gene transactivation as a transcription coactivator and protein function via poly(ADP-ribosyl)ation [1]. PARP activation is important for DNA repair, but its excessive activation is important in necrotic cell death [2]. The necrotic cell death is caused by NAD+-dependent poly(ADP-ribosyl) ation that leads to ATP depletion and metabolic collapse [23]. Prior observations by ourselves and others demonstrated that pharmacological or genetic inhibition of PARP is renoprotective against ischemia reperfusion injury [45], cisplatin nephrotoxicity [67], and obstructive nephropathy [8]. The kidney proximal tubule among renal tubules is most sensitive to lethal injury as a result of a difference in their capacity to generate energy by glycolysis [9]. The proximal tubule has a low capacity for glycolysis, as demonstrated by the failure to produce lactate under control conditions or during loss of oxidative phosphorylation using antimycin A [10]. Furthermore, the activity of hexokinase as a glycolytic enzyme is less in the proximal tubule than in the other tubules [11].
Glycolysis is the sequence of reactions that metabolizes one molecule of glucose to two molecules of pyruvate. During glycolytic flux, two molecules of ATP and two molecule of NADH are produced under an anaerobic condition. Phosphofructokinase 1 (PFK1) is one of the most important regulatory enzymes in the mammalian glycolytic pathway [12]. Phosphorylation of fructose 6-phosphate to fructose 1,6-biphosphate by PFK1 is the first point of commitment of glucose to the glycolytic pathway [13]. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is also one of the most important regulatory enzymes in glycolysis and gluconeogenesis [14]. GAPDH reversibly catalyzes the oxidation and phosphorylation of glyceraldehyde-3-phosphate to 1,3-diphosphoglycerate [14]. Poly(ADP-ribosyl)ation induced by PARP activation inhibits PFK1 and GAPDH activity in brain-derived and endothelial cells, respectively [1516]. However, the glycolytic enzymes mediated by PARP during glycolytic flux in kidney proximal tubule epithelial cells have not been identified. Here, we investigated the effect of treatment with a PARP inhibitor during glycolytic flux in kidney proximal tubule epithelial cells.
LLC-PK1 porcine kidney proximal tubule epithelial cell line was obtained from the American Type Culture Collection (Rockville, MD, USA). The cells were maintained in Dulbecco's modified Eagle's medium (DMEM)/high-glucose medium containing 10% fetal bovine serum (FBS) at 37℃ in an atmosphere of 5% CO2. The cells were grown until 70% confluence and then shifted to glucose- and serum-free DMEM medium. After treatment with 0.01, 0.1, or 1 mM 3-aminobenzamide (3-AB; R&D Systems, Minneapolis, MN, USA) in the glucose- and serum-free DMEM medium (vehicle) for 30 minutes, the cells were incubated with 10 mM glucose in XF base medium (Seahorse Bioscience, Billerica, MA, USA) with 4 mM glutamine for 30 minutes.
PARP activity in LLC-PK1 cells was measured using a universal PARP assay kit according to the manufacturer's instructions (Trevigen, Gaithersburg, MD, USA) [7]. Activities of hexokinase, phosphoglucose isomerase (PGI), GAPDH, enolase, and pyruvate kinase in the cells were measured using respective colorimetric assay kits purchased from BioVision Inc. (Mountain View, CA, USA) according to the manufacturer's instructions. PFK1 activity was measured as previously described [17]. Briefly, the cells were homogenized in cold sucrose buffer (0.32 M sucrose and 10 mM Tris-HCl, pH 7.4). Homogenates were centrifuged at 13,000 rpm for 20 minutes. The supernatants were incubated in 50 mM Tris-HCl buffer (pH 8.0) including 2.6 mM dithiothreitol, 2 mM MgCl2, 5 mM (NH4)2SO4, 1 mM EDTA, 40 units aldose, 250 units triosephosphate isomerase, 40 units α-glycerophosphate dehydrogenase, 100 mM fructose-6-phosphate, 100 mM ATP, and 16 mM NADH. The decrease in optical density at 340 nm due to the oxidation of NADH was the measured for 60 seconds.
We performed electrophoresis of protein extracts using trisglycine buffer systems and subsequent blotting as previously described [18]. Membranes were incubated with antibodies against PARP (catalog No. 13371-1-AP), hexokinase (catalog No. 19662-1-AP), PFK1 (catalog No. 55028-1-AP), and GAPDH (catalog No. 10494-1-AP) purchased from Proteintech (Chicago, IL, USA). Peroxidase-conjugated secondary antibodies (Vector Laboratories, Burlingame, CA, USA) were applied, and a chemiluminescence reagent (PerkinElmer, Boston, MA, USA) was used to detect proteins. Anti-β-actin antibody (Sigma, St. Louis, MO, USA) was used for loading controls on stripped membranes. The bands were quantified using NIH ImageJ program.
Extracellular acidification rate (ECAR) in LLC-PK1 cells was measured using an XFp extracellular flux analyzer (Seahorse Bioscience). The cells were seeded in XFp cell culture miniplates (Seahorse Bioscience) at a density of 105 cells per well in DMEM/high-glucose medium containing 10% FBS and incubated overnight. The following day, the cells were treated with 1 mM 3-AB in glucose- and serum-free DMEM medium (vehicle) for 30 minutes, and then incubated at 37℃ with XF base medium containing 4 mM glutamine in a CO2-free incubator for 60 minutes. Glycolytic flux (basal glycolysis, glycolytic activity, and glycolytic capacity) as assessed by ECAR was analyzed by the sequential injection of 10 mM glucose, 1 mM oligomycin, and 50 mM 2-deoxyglucose. ECAR was measured at 37℃ with a 3-minute mix, 0-minute wait, and 3-minute measurement protocol. The levels of ECAR were determined three times in respective phases, and expressed as units of milli-pH (mpH) per minute.
LLC-PK1 cells seeded in XFp cell culture miniplates (Seahorse Bioscience) at a density of 105 cells per well in DMEM/high-glucose medium containing 10% FBS were incubated overnight. Cells were treated with 1 mM 3-AB in glucose- and serum-free DMEM medium (vehicle) for 30 minutes; changed to XF base medium containing 4 mM glutamine, 1 mM pyruvate, and 25 mM glucose; and incubated at 37℃ in a CO2-free incubator for 60 minutes. Mitochondrial function (basal respiration, mitochondrial ATP production, and maximal respiration) as assessed by oxygen consumption rate (OCR) was analyzed by the sequential injection of 1 µM oligomycin, 2 µM carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP), and 0.5 µM rotenone plus antimycin A in the XFp extracellular flux analyzer (Seahorse Bioscience). OCR was measured at 37℃ with a 3-minute mix, 0-minute wait, and 3-minute measurement protocol. OCR was determined three times in respective phases, and expressed as units of picomoles (pmol) per minute.
LLC-PK1 cells were seeded at a density of 105 cells per well on a 24-well plate, and the next day treated with or without glucose plus/minus 3-AB as indicated. Tetramethylrhodamine, ethyl ester (TMRE; 20 nM, Abcam, Cambridge, MA, USA) was added to the cells and incubated for 30 minutes. After washing three times with 500 µl of phosphate buffered saline/0.2% FBS three-times, the cells were read using a FilterMax F3 multimode microplate reader (Molecular Devices, Sunnyvale, CA, USA) at excitation and emission wavelengths of 549 and 575 nm, respectively.
PARP activity after incubation with glucose was measured in LLC-PK1 cells. Cells incubated with glucose for 30 minutes displayed a significant increase in PARP activity, compared to that in glucose-starved control cells (Fig. 1A), but not a significant alteration in PARP expression (Fig. 1B). We also tested whether treatment with 3-AB, a PARP inhibitor, reduced PARP activity increased by glucose. Treatment with 3-AB 30 minutes prior to incubation with glucose dose-dependently diminished the increment in the PARP activity after 30 minutes of incubation with glucose (Fig. 1A). However, treatment with 3-AB in glucose-starved control cells did not significantly alter the activity of PARP protein (Fig. 1A). In addition, since that 3-AB inhibits PARP activation by competitively interfering with the binding of NAD to its active site, no alteration of PARP expression by treatment with 3-AB was confirmed. These data indicate that PARP inhibition is efficacious against PARP activation induced by glucose in kidney proximal tubule epithelial cells.
To analyze the effect of glucose-induced PARP activation on glycolysis in kidney proximal tubule epithelial cells, we conducted a real-time analysis of glycolytic flux using XFp extracellular flux analysis in LLC-PK1 cells treated with vehicle and 3-AB. No significant difference in basal glycolysis as a basal ECAR rate reached by the cells during glucose starvation was found in cells treated with vehicle and 3-AB (Fig. 2A, C). To measure glycolytic activity, glucose was injected into culture wells. ECAR was increased by glucose in cells treated with vehicle, and the level was increased further in cells treated with 3-AB (Fig. 2A). The results indicated that glycolytic activity in cells treated with 3-AB is greater than that in cells treated with vehicle (Fig. 2D). To measure glycolytic capacity as a maximum ECAR rate reached by the cells, then oligomycin was injected into culture wells. Oligomycin increased ECAR levels in cells treated with vehicle and 3-AB, resulting in no significant difference of glycolytic capacity (Fig. 2A, E). These data indicate that PARP inhibition increases glycolytic activity in kidney proximal tubule epithelial cells.
To determine whether PARP activation causes mitochondrial dysfunction in kidney proximal tubule epithelial cells, OCR was monitored by XFp extracellular flux analysis in LLC-PK1 cells. No significant difference in basal respiration as an energetic demand of the cells under the baseline condition, mitochondrial ATP production as a FCCP-sensitive OCR rate, and maximal respiration as a maximum OCR rate of respiration was found in cells treated with vehicle and 3-AB (Fig. 3A–E). Since the opening of mitochondrial permeability transition pore can precipitate the cessation of ATP synthesis in mitochondria, we assessed mitochondrial membrane potential in the cells using TMRE assay. Consistent with mitochondrial ATP production, mitochondrial membrane potential was not significantly altered by treatment with 3-AB in the cells treated with or without glucose (Fig. 4). These data suggest that PARP activation did not contribute to mitochondrial function in kidney proximal tubule epithelial cells.
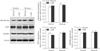
To clarify the effect of PARP inhibition on glycolysis in kidney proximal tubule epithelial cells, activities of glycolytic enzymes were measured in LLC-PK1 cells. Glucose increased the activities of glycolytic enzymes including hexokinase, PGI, PFK1, GAPDH, enolase, and pyruvate kinase, compared to the activities in glucose-starved control cells (Fig. 5A–F). Treatment with 3-AB in cells incubated with glucose markedly increased further the levels of activities in hexokinase, PFK1, and GAPDH (Fig. 5A, C, D). However, the enzyme activities were not significantly changed in glucose-starved control cells after 30 minutes of treatment with 3-AB (Fig. 5A–F). In addition, we measured expressions of those enzymes in the cells using Western blot analysis. Those enzyme expressions were not significantly altered by treatment with 3-AB in the cells treated with or without glucose (Fig. 6), indicating that the transactivation of those enzymes is independent of glucose-related metabolism and PARP inhibition. Thus, PARP activation evidently regulates hexokinase, PFK1, and GAPDH activities increased by glucose in kidney proximal tubule epithelial cells.
The present data demonstrate that PARP activation induced by glucose does not affect mitochondrial function in kidney proximal tubule epithelial cells. Instead, PARP activation leads to inhibition of glycolytic activity as determined by the significant increment induced by the injection of glucose during glycolytic flux. The inhibition of glycolytic activity is caused by the significant decrement of activity in glycolytic enzymes including hexokinase, PFK1, and GAPDH. The activation of PARP involves poly(ADP-ribose) polymerization because PARP forms homopolymers of ADP-ribose on various nuclear proteins as well as PARP itself [1]. The poly(ADP-ribosylated) proteins lose their affinity for DNA following genotoxic injury and then the proteins are inactivated [19]. In the metabolic pathway of glycolysis, hexokinase, PFK1, and GAPDH contain a poly(ADP-ribose)-binding domain including a poly(ADP-ribose)-binding motif, poly(ADP-ribose)-binding zinc finger domain, macro domain, and a domain with conservative multiple sequence alignment of two tryptophan residues and a glutamate residues [1520]. Hexokinase is the first regulatory enzyme to initiate glycolysis by converting glucose to glucose-6-phosphate [21], and its activity is inhibited by PARP activation in primary mouse cortical neurons [22]. In support of this notion, hexokinase contains a poly(ADP-ribose)-binding motif and coimmunoprecipitates with poly(ADP-ribose) after PARP activation, indicating that it is a poly(ADP-ribose)-binding protein [22]. PFK1, one of the most important regulatory enzymes of glycolysis, contains a poly(ADP-ribose)-binding domain [12]. PFK1 activity increases when the ratio of ATP to AMP is lowered [12]. The activity is inhibited by poly(ADP-ribosyl)ation induced by PARP activation in brain-derived cells [15]. The phosphorylation of fructose 6-phosphate to fructose 1, 6-bisphosphate by PFK1 is the first point of commitment of glucose to the glycolytic pathway [23]; and because this reaction involves the hydrolysis of ATP, it is essentially irreversible. Importantly, PFK1 activity is rate-limiting and therefore may be critical in determining the glycolytic activity. GAPDH is also a key enzyme in the glycolytic pathway and has a susceptibility to oxidative modifications of thiols that inhibits its activity [24]. GAPDH activity is also inhibited by poly(ADP-ribosyl)ation in kidney proximal tubule epithelial cells after ischemia reperfusion injury [25]. Poly(ADP-ribose) is detected in GAPDH, and then its activity is subsequently decreased [15]. The previous findings are consistent with our present results, suggesting that PARP activation induced by injecting glucose into kidney proximal tubule epithelial cells generates poly(ADP-ribose) on its binding site in hexokinase, PFK1, and GAPDH, reducing their activities. The poly(ADP-ribose)-binding domain in other glycolytic enzymes including PGI, aldoase, triose phosphate isomerase, phosphoglycerate kinase, phosphoglyceromutase, enolase, and pyruvate kinase has not been reported. Presently, the activities of PGI, enolase, and pyruvate kinase were not consistently altered by PARP inhibition, indicating that these enzymes may not contain the poly(ADP-ribose)-binding domain. Because of the PARP-independent enzymes including PGI, enolase, and pyruvate kinase; oligomycin-induced glycolytic capacity revealing the maximum of glycolysis in the cells might not be different between vehicle- and 3-AB–treated cells.
In glucose metabolism, glycolysis is the metabolic pathway that converts glucose into pyruvate in the cytoplasm, which produces ATP [26]. Exogenous glucose, the most important energy-producing molecule of organisms, strictly induces activities of glycolytic enzymes in the entire 10-step glycolysis pathway, with each chemical reaction catalyzed by a specific enzyme [27]. PARP inhibits glycolysis in kidneys after ischemia reperfusion injury, as demonstrated by lactate production increased by PARP deficiency in injured tissues [25]. The present data using XFp extracellular flux analysis shows that glucose increases glycolytic activity during glycolytic flux in kidney proximal tubule epithelial cells. Furthermore, treatment with the PARP inhibitor 3-AB in those cells markedly elevates glycolytic activity induced by glucose. Intriguingly, the mitochondrial basal respiration, mitochondrial ATP production, and maximal respiration were not significantly different in cells treated with 3-AB and vehicle. Our result contrasts with the previous demonstration of increased mitochondrial function through SIRT1 in PARP-deficient mice [28]. The previous report focused on the role for PARP in oxidative metabolism through SIRT1 modulation under diet-induced obesity. Similarly, high dose of glucose (30 mM) induces oxidative stress and DNA damage through SIRT1 modulation in hepatocytes, resulting in glucose toxicity [29]. In our study, the incubation with low dose of glucose (10 mM) may cause no effect on mitochondrial function. Furthermore, because myoblast and hepatocyte are more susceptible to glucose-related toxicity and metabolism compared to other epithelial cells including kidney tubular cells [30313233], the alteration of mitochondrial function by PARP activation may be dependent on cell type or tissue specific. Taken together, the present results demonstrate that exogenous glucose increases PARP activation in kidney proximal tubule epithelial cells, and that PARP activation regulates glycolytic activity through poly(ADP-ribosyl)ation of hexokinase, PFK1, and GAPDH. PARP may be a pivotal molecule involved in regulation of glucose metabolism.
Figures and Tables
Fig. 1
Treatment with 3-aminobenzamide (3-AB) attenuates poly(ADP-ribose) polymerase (PARP) activation increased by glucose in LLC-PK1 kidney proximal tubule epithelial cells. (A) PARP activity in the cells measured using the universal PARP assay kit was expressed as units (U) per mg protein. (B) PARP expression was examined by Western blot analysis using anti-PARP1 antibody. Anti–β-actin antibody was used as a loading control. Error bars represent SD (n=4 experiments). a)P<0.05 vs. control. b)P<0.05 vs. vehicle.

Fig. 2
Poly(ADP-ribose) polymerase inhibition augments glycolytic activity in LLC-PK1 kidney proximal tubule epithelial cells using XFp extracellular flux analysis. (A) Extracellular acidification rate (ECAR) analysis in 3-aminobenzamide (3-AB)–treated cells. (B) Profile of ECAR analysis. (C) Basal glycolysis indicates a basal ECAR rate reached by the cells during glucose starvation. It was calculated by the average of three ECAR baselines before glucose injection minus the average of three non-glycolytic ECAR levels after 2-deoxyglucose (2-DG) injection. (D) Glycolytic activity indicates an ECAR rate reached by the cells after the injection of saturating amounts of glucose. It was calculated by the average of three ECAR levels after glucose injection minus the average of three ECAR baselines. (E) Glycolytic capacity indicates a maximum ECAR rate reached by the cells. It was calculated by the average of three ECAR levels after oligomycin injection minus the average of three non-glycolytic ECAR levels after 2-DG injection. Error bars represent SD (n=4 experiments). a)P<0.05 vs. vehicle. BG, basal glycolysis; G, glucose; GA, glycolytic activity; GC, glycolytic capacity; O, oligomycin.
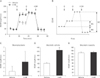
Fig. 3
Poly(ADP-ribose) polymerase is not involved in respiration and ATP production in mitochondria during treatment with glucose in LLC-PK1 kidney proximal tubule epithelial cells using XFp extracellular flux analysis. (A) Oxygen consumption rate (OCR) analysis in 3-aminobenzamide (3-AB)–treated LLC-PK1 cells. (B) Profile of OCR analysis. (C) Basal respiration indicates an energetic demand of the cells under the baseline condition. It was calculated by the average of three OCR baseline measurements before oligomycin injection minus the average of three OCR levels after rotenone and antimycin A (R&A) injection. (D) Mitochondrial ATP indicates a level of ATP produced by mitochondria. It was calculated by the average of three OCR baseline determinations before oligomycin injection minus the average of three extracellular acidification rate levels after oligomycin injection. (E) Maximal respiration indicates a maximum OCR rate of respiration that the cells can achieve. It was calculated by the average of three OCR levels after FCCP injection minus the average of three OCR levels after R&A injection. Error bars represent SD (n=4 experiments). BR, basal respiration; F, carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP); MA, mitochondrial ATP; MR, maximal respiration; O, oligomycin.
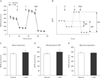
Fig. 4
Poly(ADP-ribose) polymerase and glucose-related metabolism are not involved in mitochondrial membrane potential in LLC-PK1 kidney proximal tubule epithelial cells. LLC-PK1 cells on an 24-well plate were treated with 1 mM 3-aminobenzamide (3-AB) in glucose- and serum-free Dulbecco's modified Eagle's medium medium (vehicle) for 30 minutes and then incubated with 10 mM glucose in XF base medium with 4 mM glutamine (control) for 30 minutes. The cells were stained with 20 nM of tetramethylrhodamine, ethyl ester for 30 minutes, washed three times with 500 µl of phosphate buffered saline/0.2% fetal bovine serum three times, and read by a FilterMax F3 multimode microplate reader (Molecular Devices, Sunnyvale, CA, USA) at excitation and emission wavelengths of 549 and 575 nm, respectively. There are no significant differences (n=4 experiments).

Fig. 5
Poly(ADP-ribose) polymerase inhibition augments activities of glycolytic enzymes increased by glucose in LLC-PK1 kidney proximal tubule epithelial cells. (A–F) Activities of hexokinase, phosphoglucose isomerase (PGI), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), enolase, and pyruvate kinase in the cells were measured by hexokinase, PGI, GAPDH, enolase, and pyruvate kinase colorimetric assay kits, respectively. (C) Phosphofructokinase 1 (PFK1) activity in the cells was measured as previously described [17]. All results were expressed as units (U) per mg protein per minutes. Error bars represent SD (n=4 experiments). a)P<0.05 vs. control. b)P<0.05 vs. vehicle. 3-AB, 3-aminobenzamide.

Fig. 6
Poly(ADP-ribose) polymerase and glucose-related metabolism are not involved in expressions of glycolytic enzymes in LLC-PK1 kidney proximal tubule epithelial cells. Hexokinase, phosphofructokinase 1 (PFK1) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expressions were examined by Western blot analysis. Anti–β-actin antibody was used as a loading control. There are no significant differences (n=4 experiments). 3-AB, 3-aminobenzamide.
References
1. Kraus WL, Lis JT. PARP goes transcription. Cell. 2003; 113:677–683.
2. Ha HC, Snyder SH. Poly(ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proc Natl Acad Sci U S A. 1999; 96:13978–13982.
3. Berger NA, Berger SJ. Metabolic consequences of DNA damage: the role of poly (ADP-ribose) polymerase as mediator of the suicide response. Basic Life Sci. 1986; 38:357–363.
4. Martin DR, Lewington AJ, Hammerman MR, Padanilam BJ. Inhibition of poly(ADP-ribose) polymerase attenuates ischemic renal injury in rats. Am J Physiol Regul Integr Comp Physiol. 2000; 279:R1834–R1840.
5. Yoon SP, Kim J. Poly(ADP-ribose) polymerase 1 activation links ischemic acute kidney injury to interstitial fibrosis. J Physiol Sci. 2015; 65:105–111.
6. Kim J, Long KE, Tang K, Padanilam BJ. Poly(ADP-ribose) polymerase 1 activation is required for cisplatin nephrotoxicity. Kidney Int. 2012; 82:193–203.
7. Park S, Yoon SP, Kim J. Cisplatin induces primary necrosis through poly(ADP-ribose) polymerase 1 activation in kidney proximal tubular cells. Anat Cell Biol. 2015; 48:66–74.
8. Kim J, Padanilam BJ. Loss of poly(ADP-ribose) polymerase 1 attenuates renal fibrosis and inflammation during unilateral ureteral obstruction. Am J Physiol Renal Physiol. 2011; 301:F450–F459.
9. Lieberthal W, Nigam SK. Acute renal failure. I. Relative importance of proximal vs. distal tubular injury. Am J Physiol. 1998; 275(5 Pt 2):F623–F631.
10. Bagnasco S, Good D, Balaban R, Burg M. Lactate production in isolated segments of the rat nephron. Am J Physiol. 1985; 248(4 Pt 2):F522–F526.
11. Vandewalle A, Wirthensohn G, Heidrich HG, Guder WG. Distribution of hexokinase and phosphoenolpyruvate carboxykinase along the rabbit nephron. Am J Physiol. 1981; 240:F492–F500.
12. Chesney J. 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase and tumor cell glycolysis. Curr Opin Clin Nutr Metab Care. 2006; 9:535–539.
13. Mor I, Cheung EC, Vousden KH. Control of glycolysis through regulation of PFK1: old friends and recent additions. Cold Spring Harb Symp Quant Biol. 2011; 76:211–216.
14. Chuang DM, Hough C, Senatorov VV. Glyceraldehyde-3-phosphate dehydrogenase, apoptosis, and neurodegenerative diseases. Annu Rev Pharmacol Toxicol. 2005; 45:269–290.
15. Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, Poirier GG. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008; 36:6959–6976.
16. Du X, Matsumura T, Edelstein D, Rossetti L, Zsengellér Z, Szabó C, Brownlee M. Inhibition of GAPDH activity by poly (ADP-ribose) polymerase activates three major pathways of hyperglycemic damage in endothelial cells. J Clin Invest. 2003; 112:1049–1057.
17. Kim J, Devalaraja-Narashimha K, Padanilam BJ. TIGAR regulates glycolysis in ischemic kidney proximal tubules. Am J Physiol Renal Physiol. 2015; 308:F298–F308.
18. Lee JS, Lim JY, Kim J. Mechanical stretch induces angiotensinogen expression through PARP1 activation in kidney proximal tubular cells. In Vitro Cell Dev Biol Anim. 2015; 51:72–78.
19. Althaus FR, Richter C. ADP-ribosylation of proteins: enzymology and biological significance. Mol Biol Biochem Biophys. 1987; 37:1–237.
20. Krietsch J, Rouleau M, Pic É, Ethier C, Dawson TM, Dawson VL, Masson JY, Poirier GG, Gagné JP. Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol Aspects Med. 2013; 34:1066–1087.
21. Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011; 27:441–464.
22. Andrabi SA, Umanah GK, Chang C, Stevens DA, Karuppagounder SS, Gagné JP, Poirier GG, Dawson VL, Dawson TM. Poly(ADP-ribose) polymerase-dependent energy depletion occurs through inhibition of glycolysis. Proc Natl Acad Sci U S A. 2014; 111:10209–10214.
23. Weber G. Enzymology of cancer cells (second of two parts). N Engl J Med. 1977; 296:541–551.
24. Mohr S, Stamler JS, Brüne B. Mechanism of covalent modification of glyceraldehyde-3-phosphate dehydrogenase at its active site thiol by nitric oxide, peroxynitrite and related nitrosating agents. FEBS Lett. 1994; 348:223–227.
25. Devalaraja-Narashimha K, Padanilam BJ. PARP-1 inhibits glycolysis in ischemic kidneys. J Am Soc Nephrol. 2009; 20:95–103.
26. Zwerschke W, Mazurek S, Stöckl P, Hütter E, Eigenbrodt E, Jansen-Dürr P. Metabolic analysis of senescent human fibroblasts reveals a role for AMP in cellular senescence. Biochem J. 2003; 376(Pt 2):403–411.
27. Greiner EF, Guppy M, Brand K. Glucose is essential for proliferation and the glycolytic enzyme induction that provokes a transition to glycolytic energy production. J Biol Chem. 1994; 269:31484–31490.
28. Bai P, Cantó C, Oudart H, Brunyánszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, Schoonjans K, Schreiber V, Sauve AA, Menissier-de Murcia J, Auwerx J. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 2011; 13:461–468.
29. Pang J, Gong H, Xi C, Fan W, Dai Y, Zhang TM. Poly(ADP-ribose) polymerase 1 is involved in glucose toxicity through SIRT1 modulation in HepG2 hepatocytes. J Cell Biochem. 2011; 112:299–306.
30. Zhao Y, Butler EB, Tan M. Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis. 2013; 4:e532.
31. Dhanya R, Arun KB, Syama HP, Nisha P, Sundaresan A, Santhosh Kumar TR, Jayamurthy P. Rutin and quercetin enhance glucose uptake in L6 myotubes under oxidative stress induced by tertiary butyl hydrogen peroxide. Food Chem. 2014; 158:546–554.
32. Petrescu AD, McIntosh AL, Storey SM, Huang H, Martin GG, Landrock D, Kier AB, Schroeder F. High glucose potentiates L-FABP mediated fibrate induction of PPARalpha in mouse hepatocytes. Biochim Biophys Acta. 2013; 1831:1412–1425.
33. DeFronzo RA, Davidson JA, Del Prato S. The role of the kidneys in glucose homeostasis: a new path towards normalizing glycaemia. Diabetes Obes Metab. 2012; 14:5–14.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download