Abstract
Purpose
To analyze the feasibility of three-dimensional (3D) diffusion-weighted (DW) PSIF (reversed FISP [fast imaging with steady-state free precession]) sequence in order to evaluate peripheral nerves in the elbow.
Materials and Methods
Ten normal, asymptomatic volunteers were enrolled (6 men, 4 women, mean age 27.9 years). The following sequences of magnetic resonance images (MRI) of the elbow were obtained using a 3.0-T machine: 3D DW PSIF, 3D T2 SPACE (sampling perfection with application optimized contrasts using different flip angle evolution) with SPAIR (spectral adiabatic inversion recovery) and 2D T2 TSE (turbo spin echo) with modified Dixon (m-Dixon) sequence. Two observers used a 5-point grading system to analyze the image quality of the ulnar, median, and radial nerves. The signal-to-noise ratio (SNR) and contrast-to-noise ratio (CNR) of each nerve were measured. We compared 3D DW PSIF images with other sequences using the Wilcoxon-signed rank test and Friedman test. Inter-observer agreement was measured using intraclass correlation coefficient (ICC) analysis.
Results
The mean 5-point scores of radial, median, and ulnar nerves in 3D DW PSIF (3.9/4.2/4.5, respectively) were higher than those in 3D T2 SPACE SPAIR (1.9/2.8/2.8) and 2D T2 TSE m-Dixon (1.7/2.8/2.9) sequences (P < 0.05). The mean SNR in 3D DW PSIF was lower than 3D T2 SPACE SPAIR, but there was no difference between 3D DW PSIF and 2D T2 TSE m-Dixon in all of the three nerves. The mean CNR in 3D DW PSIF was lower than 3D T2 SPACE SPAIR and 2D T2 TSE m-Dixon in the median and ulnar nerves, but no difference among the three sequences in the radial nerve.
In the case of peripheral neuropathies, traditional clinical examinations relied upon electrodiagnostic testing. However, magnetic resonance imaging (MRI) of nerves, magnetic resonance neurography (MRN), is used increasingly recently. MRN is able to define peripheral nerve anatomy and pathology (1). It has been used to confirm clinical suspicion of peripheral neuropathy by directly showing the pathologic lesion, such as nerve abnormalities or regional muscle denervation, as well as the lesions that cause nerve entrapment or impingement.
These advantages have been achieved from advancements in MRI techniques, such as high-quality three-dimensional (3D) imaging, 3.0T MRI systems, as well as new MRN sequences (2345).
In MRN, the high quality of the 2D imaging with both non-fat-suppressed T1-weighted and fat suppressed (FS) fluid-sensitive T2 weighted images (WI) is essential to evaluate different tissues, such as various intraneural and perineural anatomic and pathologic characteristics (1). Although 2D imaging remains the standard for the initial examination, the high quality 3D imaging is essential for excellent depiction of 3D nerve anatomy and pathology. Three-dimensional imaging also provides views from various planes, allowing a better understanding of disease process and localization to referring physicians (3).
Among the 3D imaging techniques, FS T2 SPACE (sampling perfection with application optimized contrasts using different flip angle evolution, Siemens Health Care, Erlangen, Germany) has been commonly used for MRN lately (3). In recent studies, many authors used FS techniques with T2 SPACE, such as 3D short tau inversion recovery (STIR) SPACE for plexus imaging and 3D spectral adiabatic inversion recovery (SPAIR) SPACE for extremities due to higher signal-to-noise ratio (SNR) (3). These sequences allow curved and/or multiplanar reconstruction. They also allowed for the creation of maximum intensity projection images of nerves for an excellent display of normal and abnormal nerves along their long axes. However, these sequences also have disadvantages due to nerve signal contamination by vessel signals. This contamination is caused by similar signal hyperintensities between the nerves and vessels. It results in grainy and noisy changes if the imaging is not properly performed (4).
Therefore, recent studies have used fat-suppressed 3D isotropic diffusion-weighted (DW) PSIF (reversed FISP [fast imaging with steady-state free precession]) sequences to acquire vessel-suppressed and nerve-selective images in a 3D isotropic fashion for peripheral nerve evaluation (456).
However, there are still few published articles addressing the feasibility of fat-suppressed 3D DW PSIF sequence application for peripheral nerves, such as those in the elbow joint. Therefore, the purpose of this article is to analyze the feasibility of fat-suppressed 3D DW PSIF sequences in the evaluation of peripheral nerves of the elbow.
This prospective study was approved by the institutional review board of our hospital. Informed consent was obtained from all subjects prior to MRI examination. In June 2012, 10 asymptomatic healthy volunteers were enrolled (6 men, 4 women, mean age 27.9 years. [range, 23-41 years]). These volunteers underwent MRI examinations of the elbow.
MR neurography was performed with a clinical 3.0-T MRI system (Siemens Magnetom Skyra 3T system, Siemens Medical Solutions, Erlangen, Germany) using a 4-channel large flex coil at the elbow joint. Images were obtained by using three imaging protocols from any side of the elbow joint. The imaging protocols included fat-suppressed 3D DW PSIF, 3D isotropic T2 SPACE with SPAIR, and 2D TSE (turbo spin echo) T2 with modified Dixon sequences. Magnetic resonance sequence parameters are listed in Table 1.
For each nerve, image quality was visually graded using a 5-point scale, modified from 3-point scales used by other researchers to score the peripheral nerves in MR imaging (46). The 5-point scale was defined as follows: 5 = excellent (well demarcated nerve, well differentiated from the background structures such as adjacent vessels, fat and muscles); 4 = good (good demarcation [focal ill-defined], good differentiation between nerves and background structures); 3 = moderate (moderate demarcation [< 50% ill-defined outer margin]); 2 = poor (poorly demarcated [> 50% ill-defined outer margin]); 1: nondiagnostic or unreadable. After 3 cases review by consensus, 2 board-certified radiologists (3, 12 years of experience in musculoskeletal imaging each) independently evaluated the axial images of the ulnar, median, and radial nerves at the elbow. All the 3 sequences were evaluated simultaneously, and the evaluated axial levels were the cubital tunnel level for the ulnar nerve and the radial head level for the median and radial nerves.
Quantitative analysis was performed using the SNR and the contrast-to-noise ratio (CNR) to provide a relative comparison between the quantitative image analysis values among the three sequences in each of the three nerves. This analysis was performed by one board-certified radiologist (12 years of experience in musculoskeletal imaging) and one 3rd grade radiology resident. SNRs (SNRnerve = Signalnerve/noise) and CNRs (CNRnerve = [Signalnerve - Signalmatched muscle]/noise) were measured in regions-of-interest (ROIs) in the nerves in correlation to the adjacent matched muscles (radial and median nerve: brachialis muscle; ulnar nerve: flexor digitorum superficialis muscle). The noise was measured as the standard deviation (SD) of the signal in the surrounding air.
The 5-point score, SNRs, CNRs of each nerve were compared between the fat-suppressed 3D DW PSIF, 3D T2 SPACE SPAIR, and 2D T2 TSE m-Dixon images.
All data were presented in mean values with SD from the data of the board-certified radiologist (12 years of experience in musculoskeletal imaging). Intra-group analysis was done with nonparametric analysis of variance for repeated measurements (Friedman). Wilcoxon-signed rank test was used for post hoc analysis when a statistical significance was detected by the Friedman test. A P-value of 0.05 was used to indicate statistical significance.
Intraclass correlation coefficient (ICC) analysis was performed to evaluate the interobserver agreement between the two radiologists in the 5-point scale from the qualitative study and between the one board-certified radiologist (12 years of experience in musculoskeletal imaging) and one 3rd grade radiology resident in the quantitative study.
From the ten elbow MR sets of asymptomatic volunteers, we obtained qualitative analysis scores and mean values (Fig. 1). There were significant differences in the mean values of 5-point scores in each nerve between the 3D DW PSIF and 3D T2 SPACE SPAIR (P < 0.05), and between the 3D DW PSIF and 2D T2 TSE m-Dixon sequences (P < 0.05) (mean values with SD of 3D DW PSIF - radial nerve: 3.9 ± 1.20, median nerve: 4.2 ± 1.03, ulnar nerve: 4.5 ± 0.97; 3D T2 SPACE SPAIR - radial: 1.9 ± 0.57, median: 2.8 ± 0.63, ulnar: 2.8 ± 0.42; 2D T2 TSE m-Dixon - radial 1.7 ± 0.48, median: 2.8 ± 0.63, ulnar: 2.9 ± 0.57). These results demonstrate higher mean values of the 5-point score in each nerve using the 3D DW PSIF sequence than any other sequence (Fig. 2). The results showed the most noticeable difference, especially in the radial nerve (Fig. 1). The range of ICC values between the two observers was 0.077 - 0.901 (3D DW PSIF: 0.841 - 0.901; 3D T2 SPACE SPAIR: 0.077-0.467; 2D T2 TSE m-DIXON: 0.208 - 0.516).
In the quantitative analysis, the 3D DW PSIF sequence showed significantly lower SNR for each of the three nerves than the 3D T2 SPACE SPAIR sequence. However, there was no statistical difference between the values of SNR of all three nerves in the 3D DW PSIF sequence and the 2D T2 TSE m-Dixon sequence (Table 2, Fig. 3). In terms of CNR, the 3D DW PSIF sequence had significantly lower values of the median and ulnar nerve than the 3D T2 SPACE SPAIR and 2D T2 TSE m-Dixon sequences, but there were no significant differences among the three sequences in the radial nerve (Table 2, Fig. 4). The range of ICC values between the two observers was 0.125 - 0.924 (3D DW PSIF - SNR: 0.714 - 0.833, CNR: 0.413 - 0.586; 3D T2 SPACE SPAIR - SNR: 0.452 - 0.739, CNR: 0.125 - 0.596; 2D T2 TSE m-Dixon - SNR: 0.787 - 0.924, CNR: 0.661 - 0.841).
MRN has recently begun to play a role in the evaluation of peripheral nerve disease. MRN not only reveals the morphological characteristics of nerves, such as caliber, continuity, and relationships to other structures, but also provides information on pathological processes, such as nerve fibrosis, inflammation, and edema (1). Originally developed in 1992, MRN was applied to evaluate large lesions such as brachial/lumbar plexus or nerve root lesions. However, as MR technology evolves, it could also be used to differentiate smaller structures, such as peripheral nerves in the elbow, wrist or foot, from surrounding tissues (78). MRN has been performed for more than two decades, with great emphasis on 2D imaging (3).
In most radiological departments, peripheral nerves are currently evaluated by using fat-suppressed T2-weighted images (T2WI) or STIR images and 3D or thin section 2D T1-weighted images (T1WI) (1). The combination of these sequences constitutes conventional MRN. Despite high resolution, soft tissue contrast, and SNR of the abovementioned conventional MRN sequences, the similar signal intensities of nerves and vessels make it difficult for them to be differentiated (4).
Since saturation bands do not work well for obliquely coursing neurovascular bundles in the extremities, any selective nerve imaging includes some diffusion weighting. This diffusion weighting suppresses the flowing vascular signal and enhances the adjacent nerve signal within the neurovascular bundle (456). Due to the steady state nature and low diffusion moment, there is adequate suppression of the vascular signal on this sequence, creating nerve-selective images. In most cases, a diffusion moment value of 80 - 90 s/mm-2 provides an acceptable compromise in peripheral nerve-to-background contrast and image signal-to-noise ratios (45). This provides selective suppression of all moving structures, including vascular flow, thereby enabling selective visualization of the nerves (1). Therefore, the 3D DW PSIF sequences allows for fine vessel-suppressed nerve-selective images in a 3D isotropic fashion (459). As the diffusion gradient is added to the 3D imaging, however, the SNR decreases and the image quality can be degraded. Disadvantages include susceptibility to local inhomogeneity, and sensitivity to motion artifacts (4).
Nonetheless, it should be noted that this sequence cannot replace conventional MRN sequences. Initial imaging evaluations should be accomplished on conventional axial T1WI and fat-suppressed T2WI, because these have better soft tissue contrast and SNR than other sequences (35).
In recent studies, FS 3D DW PSIF sequence was applied to define peripheral nerves and to visualize diffuse/focal peripheral neuropathy at the knee, calf, foot, elbow, and wrist (510). These studies revealed that the fat-suppressed 3D DW PSIF sequence improved identification of peripheral nerves compared to that using T2WI (4).
There have been, however, no prior studies comparing the fat-suppressed 3D DW PSIF sequence to other commonly used sequences. Therefore, this study planned to address the imaging feasibility and to compare the fat-suppressed 3D DW PSIF sequence with the 3D T2 SPACE SPAIR and 2D T2 TSE m-Dixon techniques at the peripheral nerves of the elbow. We performed MRN at the elbow joint to evaluate commonly affected peripheral nerves, and to obtain various peripheral nerve images concurrently. Our results from asymptomatic volunteers demonstrate that there were significant differences in the nerve image qualities evaluated by the 5-point score between the fat-suppressed 3D DW PSIF sequence and the two other sequences. This finding may have resulted from the selective suppression of vascular flow signal in the 3D DW PSIF sequence. This result may suggest that the FS 3D DW PSIF sequence is superior to the others in the delineation of peripheral nerve at the elbow. However, the FS 3D DW PSIF sequence had the lower SNR and CNR values than the other sequences in general, and it was statistically significant in SNRs compared to 3D T2 SPACE SPAIR in the all 3 nerves and in CNRs compared to the 3D T2 SPACE SPAIR and the 2D T2 TSE with m-Dixon sequences in the median and ulnar nerves. This result may be due to the inherent nature of the diffusion gradient which decreases the SNR. Therefore, the fat-suppressed 3D DW PSIF sequence should be used as an additional study to the conventional sequences, such as T2 SPACE SPAIR or T2 TSE m-Dixon sequences, in order to detect the peripheral nerves easier.
The range of ICC was broad among the sequences both in the 5-point scoring system and in the measurements of SNR, and CNR. The values of ICC were relatively high in 3D DW PSIF but low in T2 SPACE SPAIR and T2 TSE m-Dixon sequence. This may be because there was difficulty in differentiating nerves from the accompanying vessels with T2 SPACE SPAIR or T2 TSE m-Dixon sequence solely without conventional T1WI or 3D DW PSIF sequence.
Our study has several limitations. First, the study population was small and not diverse. Therefore, it may not accurately represent the general population. Secondly, we compared the fat-suppressed 3D DW PSIF only with the 2 sequences, not including other more commonly used 2D T2 STIR sequence. But, the frequency of using STIR is decreasing nowadays due to its long acquisition time, and also we do not use the sequence in the routine clinical MRI for the elbow in our institute.
In conclusion, the fat-suppressed 3D DW PSIF sequence may be feasible to differentiate peripheral nerves from accompanying vascular structures of the elbow joint, despite its lower SNR and CNR values, compared to those of 3D T2 SPACE SPAIR and to 2D T2 TSE m-Dixon sequences. Future studies are needed to optimize the entire image quality and to include patients with peripheral neuropathy.
Figures and Tables
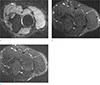 | Fig. 2MR neurography images. (a) Fat-suppressed 3D DW PSIF image, (b) Fat-suppressed 3D T2 SPAIR image, (c) Fatsuppressed 2D T2 m-Dixon image. In the fat-suppressed 3D DWI PSIF images, the median (thick arrows), radial (arrows) and ulnar nerves (curved arrows) are easily defined with suppressed adjacent vessel signals. However, in other sequences, it is difficult to differentiate the nerves from the adjacent vessel signals, even though the SNR and CNR of the nerves are better than are the fat-suppressed 3D DW PSIF images. |
Table 1
MRI Sequence Parameters

2D = two-dimensional; 3D = three-dimensional; DW = diffusion-weighted; PSIF = reversed FISP (fast imaging with steady-state free precession); SPACE = sampling perfection with application optimized contrasts using different flip angle evolution; SPAIR = spectral adiabatic inversion recovery; TSE = turbo spin echo; m-Dixon = modified Dixon; TR = repetition time; TE = echo time
Table 2
Quantitative Analysis Score of Each Volunteers

References
1. Chhabra A, Andreisek G, Soldatos T, et al. MR neurography: past, present, and future. AJR Am J Roentgenol. 2011; 197:583–591.
2. Thawait SK, Chaudhry V, Thawait GK, et al. High-resolution MR neurography of diffuse peripheral nerve lesions. AJNR Am J Neuroradiol. 2011; 32:1365–1372.
3. Chhabra A, Flammang A, Padua A Jr, Carrino JA, Andreisek G. Magnetic resonance neurography: technical considerations. Neuroimaging Clin N Am. 2014; 24:67–78.
4. Chhabra A, Soldatos T, Subhawong TK, et al. The application of three-dimensional diffusion-weighted PSIF technique in peripheral nerve imaging of the distal extremities. J Magn Reson Imaging. 2011; 34:962–967.
5. Chhabra A, Subhawong TK, Bizzell C, Flammang A, Soldatos T. 3T MR neurography using three-dimensional diffusion-weighted PSIF: technical issues and advantages. Skeletal Radiol. 2011; 40:1355–1360.
6. Zhang Z, Meng Q, Chen Y, et al. 3-T imaging of the cranial nerves using three-dimensional reversed FISP with diffusion-weighted MR sequence. J Magn Reson Imaging. 2008; 27:454–458.
7. Enochs WS, Weissleder R. MR imaging of the peripheral nervous system. J Magn Reson Imaging. 1994; 4:251–257.
8. Filler AG, Kliot M, Howe FA, et al. Application of magnetic resonance neurography in the evaluation of patients with peripheral nerve pathology. J Neurosurg. 1996; 85:299–309.
9. Chhabra A, Zhao L, Carrino JA, et al. MR neurography: advances. Radiol Res Pract. 2013; 2013:809568.
10. Umutlu L, Maderwald S, Kraff O, et al. New look at renal vasculature: 7 tesla nonenhanced T1-weighted FLASH imaging. J Magn Reson Imaging. 2012; 36:714–721.




 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download