Abstract
Objective
Pathogenesis of Behçet's disease (BD) is known to be multifactorial and accumulating data suggest genetic mechanisms. Variations in nuclear DNAs have been largely investigated, while studies on mitochondrial DNAs are limited. The purpose of the current study is to investigate associations of mitochondrial single nucleotide polymorphisms and haplotypes with BD.
Methods
Complete mitochondrial DNAs were sequenced using chip array with blood samples collected from 20 patients and 10 control subjects. Haplotypes were searched in hypervariable region 1 and 2. Chi square or Fisher's exact test was used to analyze associations of mitochondrial single nucleotide polymorphisms between two groups and associations between clinical characteristics and mitochondrial single nucleotide polymorphisms.
Results
From a total of 16,569 for each individual, 16,545 mitochondrial DNA nucleotides were sequenced. m.248A> G, m.709G> A, m.3970C> T, m.6392T> C, m.6962G> A, m.10310G> A, m.10609T> C, m.12406G> A, m.12882C> T, m.13928G> C, m.16129G> A, and m16304T> C were observed more frequently in the patient group, although without statistical significance, while m.304C> A, m.3010G> A, m.4883C> T, m.5178C> A, and m.14668C> T were more frequent in the control group (p=0.008, 0.026, 0.007, 0.007, and 0.026, respectively). m.16182A> C, m.16183A> C, and m.16189T> C were associated with uveitis (p=0.041, 0.022, and 0.014, respectively). None of the haplotypes we searched were statistically associated with BD risk, but B4a was observed more frequently in the patient group.
Conclusion
We report the first association study between BD and mitochondrial single nucleotide polymorphisms in a Korean population. In the current study, m.248A> G, m.709G> A, m.3970C> T, m.6392T> C, m.6962G> A, m.10310G> A, m.10609T> C, m.12406G> A, m.12882C> T, m.13928G> C, m.16129G> A, and m16304 T> C could be candidate mitochondrial single nucleotide polymorphisms in BD.
REFERENCES
1. Ohno S, Ohguchi M, Hirose S, Matsuda H, Wakisaka A, Aizawa M. Close association of HLA-Bw51 with Behçet's disease. Arch Ophthalmol. 1982; 100:1455–8.
2. Mendoza-Pinto C, García-Carrasco M, Jiménez-Hernández M, Jiménez Hernández C, Riebeling-Navarro C, Nava Zavala A, et al. Etiopathogenesis of Behçet's disease. Autoimmun Rev. 2010; 9:241–5.
3. Kaya Tİ. Genetics of Behçet's disease. Patholog Res Int. 2012; 2012; 912589.
4. Xavier JM, Shafiee NM, Ghaderi F, Rosa A, Abdollahi BS, Nadji A, et al. Association of mitochondrial polymorphism m.709G> A with Behçet's disease. Ann Rheum Dis. 2011; 70:1514–6.
5. International Study Group for Behçet's Disease. Criteria for diagnosis of Behçet's disease. Lancet. 1990; 335:1078–80.
6. Kussmaul L, Hirst J. The mechanism of superoxide production by NADH:ubiquinone oxidoreductase (complex I) from bovine heart mitochondria. Proc Natl Acad Sci U S A. 2006; 103:7607–12.

7. Thomas AW, Edwards A, Sherratt EJ, Majid A, Gagg J, Alcolado JC. Molecular scanning of candidate mitochondrial tRNA genes in type 2 (non-insulin dependent) diabetes mellitus. J Med Genet. 1996; 33:253–5.

8. Maassen JA, 't Hart LM, Janssen GM, Reiling E, Romijn JA, Lemkes HH. Mitochondrial diabetes and its lessons for common Type 2 diabetes. Biochem Soc Trans. 2006; 34:819–23.

10. Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011; 12:222–30.

Figure 1.
Results of part of chip array. Mitochondrial alterations with different frequencies between two groups are shown. COI: cytochrome c oxidas, HV: hypervariable region, MT: mitochondria, ND: NADH dehydrogenase, RCRS: Cambridge Reference Sequence, SNP: single nucleotide polymorphisms.
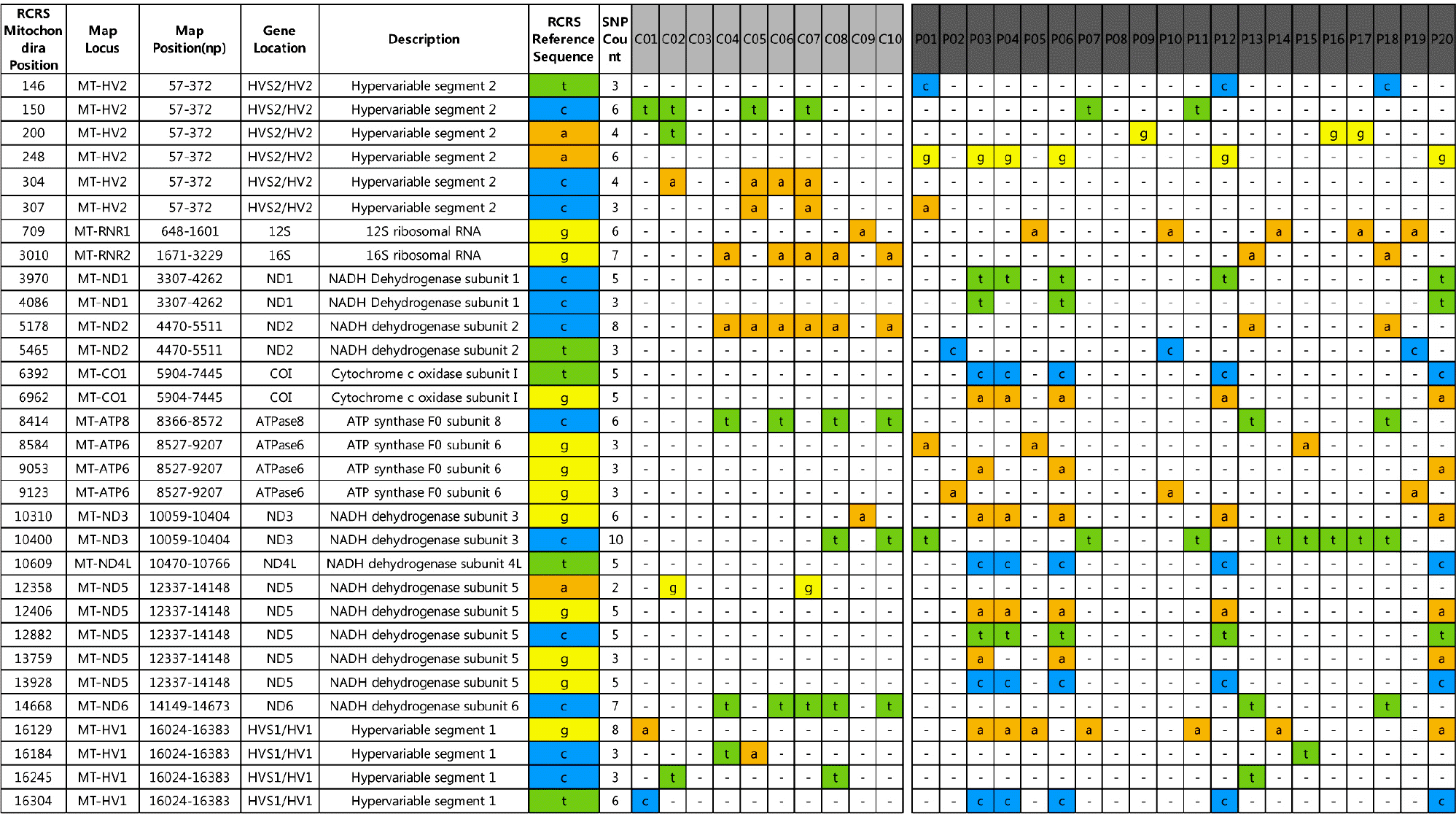
Table 1.
Clinical characteristics of patients with Behçet's disease
Table 2.
Associations between mitochondrial DNA alterations and Behçet's disease (BD)
| RCRS position | Alteration | BD (n=20) | Control (n=10) | p-value* | OR* (95% CI) |
|---|---|---|---|---|---|
| 248 | A> G | 6 (30) | 0 (0) | 0.074 | NA |
| 304 | C> A | 0 (0) | 4 (40) | 0.008 | NA |
| 709 | G> A | 5 (25) | 1 (10) | 0.633 | 3.000 (0.301∼29.940) |
| 3010 | G> A | 2 (10) | 5 (50) | 0.026 | 0.111 (0.016∼0.755) |
| 3970 | C> T | 5 (25) | 0 (0) | 0.140 | NA |
| 4883 | C> T | 2 (10) | 6 (60) | 0.007 | 0.074 (0.011∼0.512) |
| 5178 | C> A | 2 (10) | 6 (60) | 0.007 | 0.074 (0.011∼0.512) |
| 6392 | T> C | 5 (25) | 0 (0) | 0.140 | NA |
| 6962 | G> A | 5 (25) | 0 (0) | 0.140 | NA |
| 10310 | G> A | 5 (25) | 1 (10) | 0.633 | 3.000 (0.301∼29.940) |
| 10609 | T> C | 5 (25) | 0 (0) | 0.140 | NA |
| 12406 | G> A | 5 (25) | 0 (0) | 0.140 | NA |
| 12882 | C> T | 5 (25) | 0 (0) | 0.140 | NA |
| 13928 | G> C | 5 (25) | 0 (0) | 0.140 | NA |
| 14668 | C> T | 2 (10) | 5 (50) | 0.026 | 0.111 (0.016∼0.755) |
| 16129 | G> A | 7 (35) | 1 (10) | 0.210 | 4.846 (0.505∼46.492) |
| 16304 | T> C | 5 (25) | 1 (10) | 0.633 | 3.000 (0.301∼29.940) |
Table 3.
Results of haplotype analysis*




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download