Abstract
Autophagy is a principle catabolic process mediated by lysosomes in eukaryotic cells. This is an intracellular homeostatic mechanism crucial for degradation in acidic lysosomal compartments of waste components from the cytoplasm. Autophagy research was initially focused on its degradation mechanism, but focus is now shifting to its effects on immunity. It contributes to detection and removal of pathogens as well as regulation of inflammasomes and neutrophil extracellular traps. Moreover, it is pivotal in antigen presentation and immune cell maturation, survival and homeostasis. The importance of autophagic pathways in normal and dysregulated immunity has become increasingly recognized in the past several years. Dysregulation of the autophagic pathway is implicated in the pathogenesis of several rheumatic diseases. In this review, we summarize the immunological function of autophagy in innate and adaptive immunity, and the functions of autophagy in the pathogenesis of rheumatic diseases.
Go to : 
REFERENCES
2. Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007; 27:19–40.

4. Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012; 22:407–17.

5. Yang Z, Goronzy JJ, Weyand CM. Autophagy in autoimmune disease. J Mol Med (Berl). 2015; 93:707–17.

6. Zhou XJ, Zhang H. Autophagy in immunity: implications in etiology of autoimmune/autoinflammatory diseases. Autophagy. 2012; 8:1286–99.
7. Pierdominici M, Vomero M, Barbati C, Colasanti T, Maselli A, Vacirca D, et al. Role of autophagy in immunity and autoimmunity, with a special focus on systemic lupus erythematosus. FASEB J. 2012; 26:1400–12.

8. Rashid HO, Yadav RK, Kim HR, Chae HJ. ER stress: autophagy induction, inhibition and selection. Autophagy. 2015; 11:1956–77.

9. Itakura E, Mizushima N. Characterization of autophago-some formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010; 6:764–76.

10. Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010; 22:132–9.

11. Sun Q, Zhang J, Fan W, Wong KN, Ding X, Chen S, et al. The RUN domain of rubicon is important for hVps34 binding, lipid kinase inhibition, and autophagy suppression. J Biol Chem. 2011; 286:185–91.

12. Martelli AM, Chiarini F, Evangelisti C, Cappellini A, Buontempo F, Bressanin D, et al. Two hits are better than one: targeting both phosphatidylinositol 3-kinase and mammalian target of rapamycin as a therapeutic strategy for acute leukemia treatment. Oncotarget. 2012; 3:371–94.

13. Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, et al. Protection against fatal sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998; 72:8586–96.

14. Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008; 19:2092–100.

15. Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007; 282:37298–302.

16. Mizushima N, Yoshimori T, Ohsumi Y. Role of the Apg12 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2003; 35:553–61.

17. Klionsky DJ. The molecular machinery of autophagy: un-answered questions. J Cell Sci. 2005; 118:7–18. mammalian autophagy. Int J Biochem Cell Biol. 2004; 36:2503–18.

18. Satoo K, Noda NN, Kumeta H, Fujioka Y, Mizushima N, Ohsumi Y, et al. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009; 28:1341–50.

19. Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004; 36:2503–18.

20. Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011; 469:323–35.

21. Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem. 2011; 80:125–56.

22. Jäger S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004; 117:4837–48.
23. Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, et al. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol. 2010; 188:253–69.

24. Deretic V. Autophagy as an innate immunity paradigm: expanding the scope and repertoire of pattern recognition receptors. Curr Opin Immunol. 2012; 24:21–31.

25. Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005; 120:159–62.
27. Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007; 7:767–77.

28. Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and mycobacterium tuberculosis survival in infected macrophages. Cell. 2004; 119:753–66.

29. Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, et al. Autophagy defends cells against invading group A streptococcus. Science. 2004; 306:1037–40.
30. Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007; 315:1398–401.

31. Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al. Neutrophil extracellular traps kill bacteria. Science. 2004; 303:1532–5.

32. Remijsen Q, Vanden Berghe T, Wirawan E, Asselbergh B, Parthoens E, De Rycke R, et al. Neutrophil extracellular trap cell death requires both autophagy and superoxide generation. Cell Res. 2011; 21:290–304.

33. Kono H, Kimura Y, Latz E. Inflammasome activation in response to dead cells and their metabolites. Curr Opin Immunol. 2014; 30:91–8.

34. Stutz A, Golenbock DT, Latz E. Inflammasomes: too big to miss. J Clin Invest. 2009; 119:3502–11.

35. Bodemann BO, Orvedahl A, Cheng T, Ram RR, Ou YH, Formstecher E, et al. RalB and the exocyst mediate the cellular starvation response by direct activation of autophago-some assembly. Cell. 2011; 144:253–67.

36. Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endo-toxin-induced IL-1beta production. Nature. 2008; 456:264–8.
37. Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013; 368:651–62.

38. Dengjel J, Schoor O, Fischer R, Reich M, Kraus M, Müller M, et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc Natl Acad Sci U S A. 2005; 102:7922–7.

39. Crotzer VL, Blum JS. Autophagy and intracellular surveillance: modulating MHC class II antigen presentation with stress. Proc Natl Acad Sci U S A. 2005; 102:7779–80.

40. Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, Chervonsky A, et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010; 32:227–39.

41. Münz C. Antigen processing via autophagy–not only for MHC class II presentation anymore? Curr Opin Immunol. 2010; 22:89–93.

42. Bronietzki AW, Schuster M, Schmitz I. Autophagy in T-cell development, activation and differentiation. Immunol Cell Biol. 2015; 93:25–34.

43. Suda T, Takubo K, Semenza GL. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell. 2011; 9:298–310.

44. Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008; 455:396–400.

45. Aichinger M, Wu C, Nedjic J, Klein L. Macroautophagy sub-strates are loaded onto MHC class II of medullary thymic epithelial cells for central tolerance. J Exp Med. 2013; 210:287–300.

46. Parekh VV, Wu L, Boyd KL, Williams JA, Gaddy JA, Olivares-Villagómez D, et al. Impaired autophagy, defective T cell homeostasis, and a wasting syndrome in mice with a T cell-specific deletion of Vps34. J Immunol. 2013; 190:5086–101.

47. Kovacs JR, Li C, Yang Q, Li G, Garcia IG, Ju S, et al. Autophagy promotes T-cell survival through degradation of proteins of the cell death machinery. Cell Death Differ. 2012; 19:144–52.

48. McLeod IX, Zhou X, Li QJ, Wang F, He YW. The class III kinase Vps34 promotes T lymphocyte survival through regulating IL-7Rα surface expression. J Immunol. 2011; 187:5051–61.

49. Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, et al. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009; 460:108–12.

50. Arsov I, Adebayo A, Kucerova-Levisohn M, Haye J, MacNeil M, Papavasiliou FN, et al. A role for autophagic protein be-clin 1 early in lymphocyte development. J Immunol. 2011; 186:2201–9.

51. Miller BC, Zhao Z, Stephenson LM, Cadwell K, Pua HH, Lee HK, et al. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008; 4:309–14.
52. Conway KL, Kuballa P, Khor B, Zhang M, Shi HN, Virgin HW, et al. ATG5 regulates plasma cell differentiation. Autophagy. 2013; 9:528–37.

53. Pengo N, Scolari M, Oliva L, Milan E, Mainoldi F, Raimondi A, et al. Plasma cells require autophagy for sustainable immunoglobulin production. Nat Immunol. 2013; 14:298–305.

55. Caza TN, Talaber G, Perl A. Metabolic regulation of organ-elle homeostasis in lupus T cells. Clin Immunol. 2012; 144:200–13.

56. Gros F, Arnold J, Page N, Décossas M, Korganow AS, Martin T, et al. Macroautophagy is deregulated in murine and human lupus T lymphocytes. Autophagy. 2012; 8:1113–23.

57. Alessandri C, Barbati C, Vacirca D, Piscopo P, Confaloni A, Sanchez M, et al. T lymphocytes from patients with systemic lupus erythematosus are resistant to induction of autophagy. FASEB J. 2012; 26:4722–32.

58. Moulton VR, Tsokos GC. Abnormalities of T cell signaling in systemic lupus erythematosus. Arthritis Res Ther. 2011; 13:207.

59. Lee WS, Sung MS, Lee EG, Yoo HG, Cheon YH, Chae HJ, et al. A pathogenic role for ER stress-induced autophagy and ER chaperone GRP78/BiP in T lymphocyte systemic lupus erythematosus. J Leukoc Biol. 2015; 97:425–33.

60. Clarke AJ, Ellinghaus U, Cortini A, Stranks A, Simon AK, Botto M, et al. Autophagy is activated in systemic lupus erythematosus and required for plasmablast development. Ann Rheum Dis. 2015; 74:912–20.

61. International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN). Harley JB, Alarcón-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, et al. Genomewide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008; 40:204–10.

62. Orozco G, Eyre S, Hinks A, Bowes J, Morgan AW, Wilson AG, et al. Study of the common genetic background for rheumatoid arthritis and systemic lupus erythematosus. Ann Rheum Dis. 2011; 70:463–8.

63. Perl A. Emerging new pathways of pathogenesis and targets for treatment in systemic lupus erythematosus and sjogren's syndrome. Curr Opin Rheumatol. 2009; 21:443–7.

64. Ruiz-Irastorza G, Ramos-Casals M, Brito-Zeron P, Khamashta MA. Clinical efficacy and side effects of anti-malarials in systemic lupus erythematosus: a systematic review. Ann Rheum Dis. 2010; 69:20–8.

65. Page N, Gros F, Schall N, Décossas M, Bagnard D, Briand JP, et al. HSC70 blockade by the therapeutic peptide P140 affects autophagic processes and endogenous MHCII presentation in murine lupus. Ann Rheum Dis. 2011; 70:837–43.

66. McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med. 2011; 365:2205–19.

67. Shin YJ, Han SH, Kim DS, Lee GH, Yoo WH, Kang YM, et al. Autophagy induction and CHOP under-expression promotes survival of fibroblasts from rheumatoid arthritis patients under endoplasmic reticulum stress. Arthritis Res Ther. 2010; 12:R19.

68. Xu K, Xu P, Yao JF, Zhang YG, Hou WK, Lu SM. Reduced apoptosis correlates with enhanced autophagy in synovial tissues of rheumatoid arthritis. Inflamm Res. 2013; 62:229–37.

69. Kato M, Ospelt C, Gay RE, Gay S, Klein K. Dual role of autophagy in stress-induced cell death in rheumatoid arthritis synovial fibroblasts. Arthritis Rheumatol. 2014; 66:40–8.

70. Lin NY, Beyer C, Giessl A, Kireva T, Scholtysek C, Uderhardt S, et al. Autophagy regulates TNFα-mediated joint destruction in experimental arthritis. Ann Rheum Dis. 2013; 72:761–8.

71. Li RF, Chen G, Ren JG, Zhang W, Wu ZX, Liu B, et al. The adaptor protein p62 is involved in RANKL-induced autophagy and osteoclastogenesis. J Histochem Cytochem. 2014; 62:879–88.

72. DeSelm CJ, Miller BC, Zou W, Beatty WL, van Meel E, Takahata Y, et al. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev Cell. 2011; 21:966–74.

73. Ireland JM, Unanue ER. Processing of proteins in autophagy vesicles of antigen-presenting cells generates citrullinated peptides recognized by the immune system. Autophagy. 2012; 8:429–30.

74. Valesini G, Gerardi MC, Iannuccelli C, Pacucci VA, Pendolino M, Shoenfeld Y. Citrullination and autoimmunity. Autoimmun Rev. 2015; 14:490–7.

75. Sorice M, Iannuccelli C, Manganelli V, Capozzi A, Alessandri C, Lococo E, et al. Autophagy generates citrullinated peptides in human synoviocytes: a possible trigger for anticitrullinated peptide antibodies. Rheumatology (Oxford). 2016; 55:1374–85.

76. Frech T, De Domenico I, Murtaugh MA, Revelo MP, Li DY, Sawitzke AD, et al. Autophagy is a key feature in the pathogenesis of systemic sclerosis. Rheumatol Int. 2014; 34:435–9.

77. Dumit VI, Kuttner V, Kappler J, Piera-Velazquez S, Jimenez SA, Bruckner-Tuderman L, et al. Altered MCM protein levels and autophagic flux in aged and systemic sclerosis dermal fibroblasts. J Invest Dermatol. 2014; 134:2321–30.

78. Sotgia F, Martinez-Outschoorn UE, Howell A, Pestell RG, Pavlides S, Lisanti MP. Caveolin-1 and cancer metabolism in the tumor microenvironment: markers, models, and mechanisms. Annu Rev Pathol. 2012; 7:423–67.

79. Castello-Cros R, Whitaker-Menezes D, Molchansky A, Purkins G, Soslowsky LJ, Beason DP, et al. Scleroderma-like properties of skin from caveolin-1-deficient mice: implications for new treatment strategies in patients with fibrosis and systemic sclerosis. Cell Cycle. 2011; 10:2140–50.
80. Wenink MH, Santegoets KC, Butcher J, van Bon L, Lamers-Karnebeek FG, van den Berg WB, et al. Impaired dendritic cell proinflammatory cytokine production in psoriatic arthritis. Arthritis Rheum. 2011; 63:3313–22.

81. Peral de Castro C, Jones SA, Ní Cheallaigh C, Hearnden CA, Williams L, Winter J, et al. Autophagy regulates IL-23 secretion and innate T cell responses through effects on IL-1 secretion. J Immunol. 2012; 189:4144–53.

82. Ciccia F, Accardo-Palumbo A, Rizzo A, Guggino G, Raimondo S, Giardina A, et al. Evidence that autophagy, but not the unfolded protein response, regulates the expression of IL-23 in the gut of patients with ankylosing spondylitis and subclinical gut inflammation. Ann Rheum Dis. 2014; 73:1566–74.

83. Dong G, You M, Fan H, Ding L, Sun L, Hou Y. STS-1 promotes IFN-α induced autophagy by activating the JAK1-STAT1 signaling pathway in B cells. Eur J Immunol. 2015; 45:2377–88.

84. Weindel CG, Richey LJ, Bolland S, Mehta AJ, Kearney JF, Huber BT. B cell autophagy mediates TLR7-dependent autoimmunity and inflammation. Autophagy. 2015; 11:1010–24.

85. Li B, Yue Y, Dong C, Shi Y, Xiong S. Blockade of macrophage autophagy ameliorates activated lymphocytes-derived DNA induced murine lupus possibly via inhibition of proinflammatory cytokine production. Clin Exp Rheumatol. 2014; 32:705–14.
Go to : 
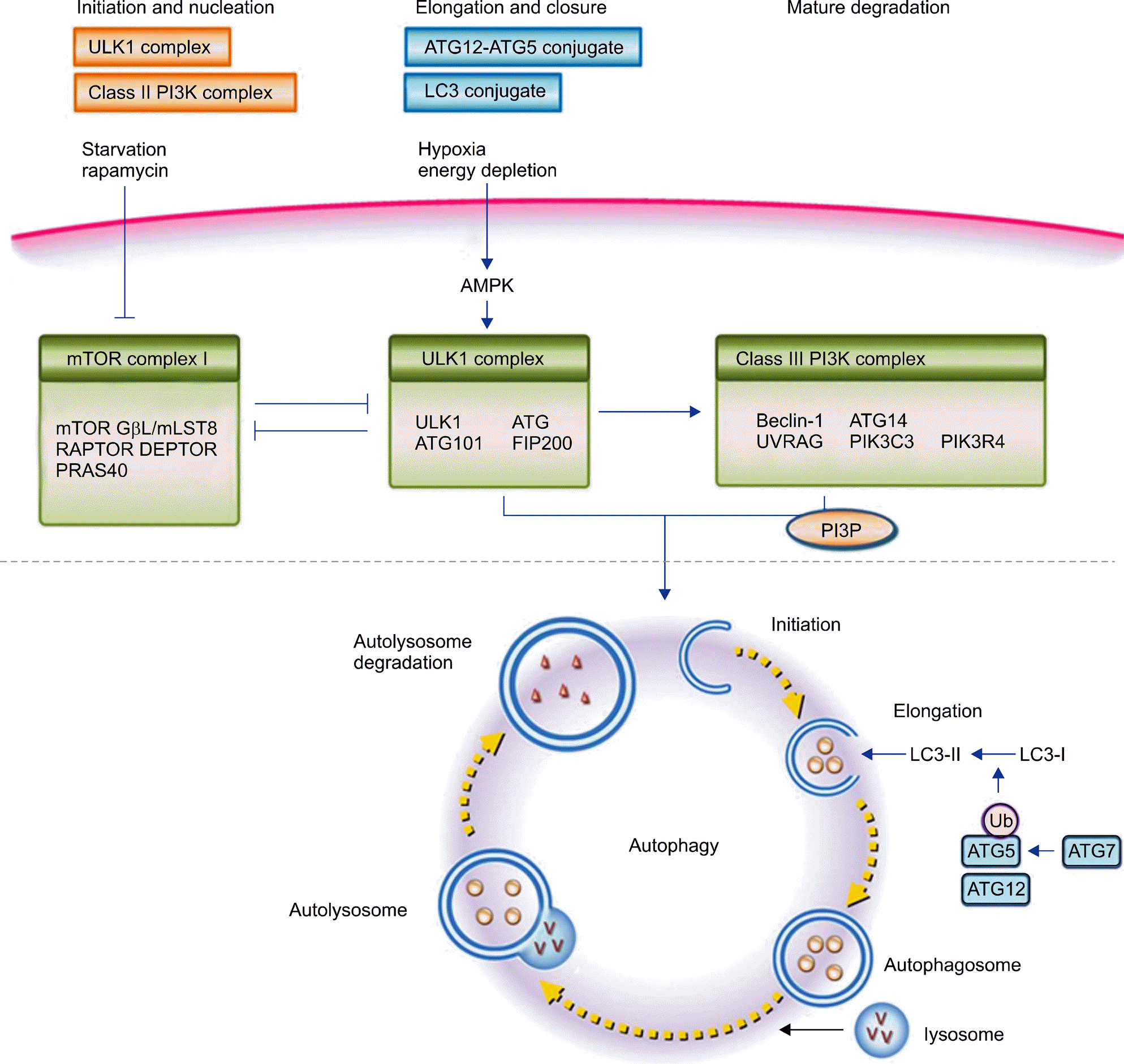 | Figure 1.Autophagic pathway. The autophagy pathway is induced by inactivation of mTOR complex. Starvation or energy ex-haustion inhibits mTOR complex, it enables ULK complex to be dissociated from mTOR complex. The stimulated ULK complex translocates to the ER, and activates class III PI3K complex. The PI3P, which is essential for autophagosomal membrane nucleation, is formed by activated class III PI3K complex. The phagophore expansion and closure mediated through conversion of LC3-I to LC3-II occurred with the help of ATG7-induced, ATG5-ATG12 complex mediated lipidation. After formation of autophagosome, it fuses with lysosome to form autolysosome and sequestered materials are degradaed by lysosomal hydrolase. Adapted from Figure 1 in the article of Levine et al. [20] (Nature 2011;469:323-35). mTOR: mammalian target of rapamycin, ULK: UNC-51-like kinase, ER: endoplasmic reticulum, PI3K: phosphatidylinositol 3-kinase, PI3P: phosphatidylinositol-3-phosphate, LC: light chain, ATG: au-tophagy-related protein, DEPTOR: DEP domain containing mTOR-interacting protein, FIP200: focal adhesion kinase family inter-acting protein of 200 kDA, Gβ L/mLST8: G protein β subunit-like protein, PRAS40: protein-rich AKT substrate 40 kDA, RAPTOR: regulatory associated protein of mTOR, PIK3C3: PI3K catalytic subunit type 3, PIK3R4: PI3K regulatory subunit 4, UVRAG: UV radiation resistance associated gene protein. |
Table 1.
Autophagic defect of immune cells in the pathogenesis of rheumatic disease
SLE: systemic lupus erythematosus, STS-1: suppressor of T-cell receptor signaling 1, JAK-STAT: Janus kinase– signal transducer and activator of transcription, IFN: interferon, Atg: autophagy-related gene, IL: interleukin, TNF-α: tumor necrosis factor-α, RA: rheumatoid arthritis, LC: light chain, ER: endoplasmic reticulum, siRNA: small interfering RNA, SSc: systemic sclerosis, PsA: psoriatic arthritis, ATG: autophagy-related protein, AS: ankylosing spondylitis, HSP: heat shock protein.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download