Abstract
Objective
Methods
Results
REFERENCES
Figure 1.
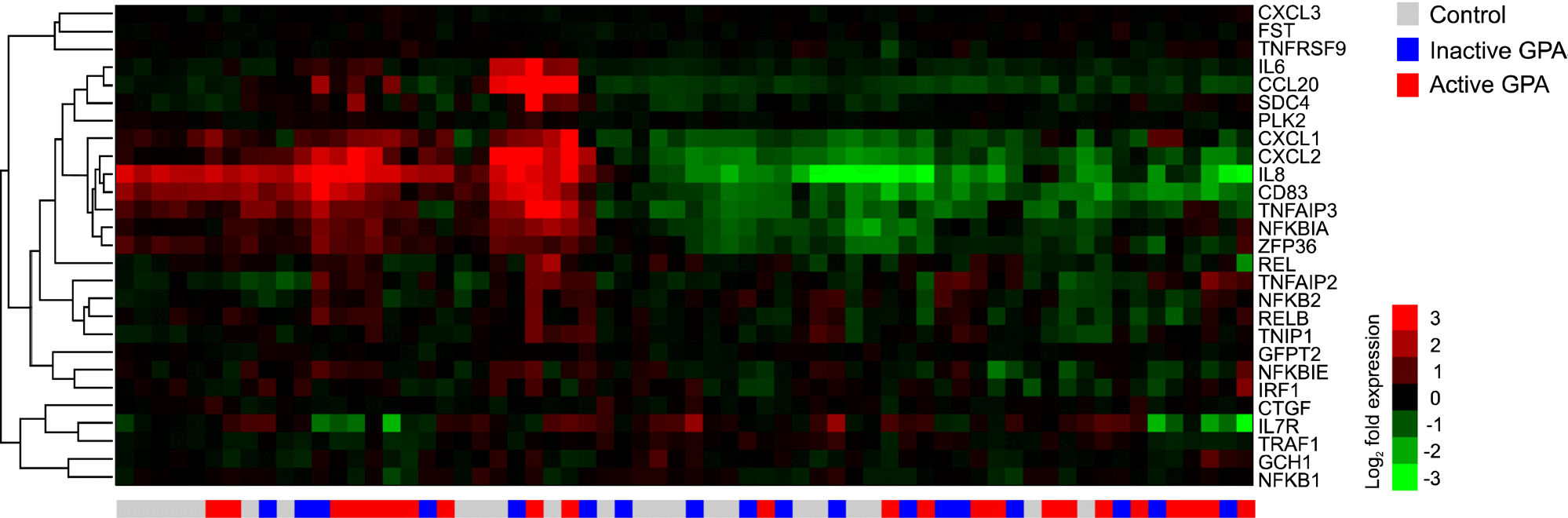
Table 1.
| Gene set name | Description* | Gene set size | NES | Nominal p | FDR† q |
|---|---|---|---|---|---|
| A. Enriched in GPA | |||||
| TIAN_TNF_SIGNALING_NOT_VIA_ NFKB | Genes modulated in HeLa cells by TNF not via NFKB pathway | 15 | 1.65 | 0.016 | 0.049* |
| ZHANG_RESPONSE_TO_IKK_INHI BITOR_AND_TNF_UP | Genes upregulated in BxPC3 cells (pancreatic cancer) after treatment with TNFIKI-1, an inhibitor of IkappaB kinase | 158 | 1.26 | 0.052 | 0.255 |
| SANA_TNF_SIGNALING_UP | Genes upregulated in five primary endothelial cell types (lung, aortic, iliac, dermal, and colon) by TNF | 55 | 1.16 | 0.229 | 0.365 |
| SANA_TNF_SIGNALING_DN | Genes downregulated in five primary endothelial cell types (lung, aortic, iliac, dermal, and colon) by TNF | 58 | 0.91 | 0.622 | 0.750 |
| TIAN_TNF_SIGNALING_VIA_NFKB | Genes modulated in HeLa cells by TNF | 19 | 0.70 | 0.884 | 0.958 |
| BIOCARTA_TNFR1_PATHWAY | TNFR1 Signaling Pathway | 28 | 0.55 | 0.980 | 1.000 |
| B. Enriched in healthy controls | |||||
| BIOCARTA_TNFR2_PATHWAY | TNFR2 Signaling Pathway | 17 | 0.65 | 0.930 | 0.991 |
GSEA showed that only one of the downstream genes of TNF-α activation was enriched in patents with GPA as compared to healthy controls. FDR: false discovery rate, GPA: granulomatosis with polyangiitis, GSEA: gene set enrichment analysis, NES: normalized enrichment score, TNF: tumor necrosis factor, TNFR: tumor necrosis fator receptor.
* Description of gene sets are available on http://www.broadinstitute.org/gsea.
Table 2.
| Gene set name | Description* | Gene set size | NES | Nominal p | FDR† q |
|---|---|---|---|---|---|
| A. Enriched in GPA | |||||
| HUTTMANN_B_CLL_POOR_SURV IVAL_UP | Up-regulated genes in B-CLL patients expressing high levels of ZAP70 and CD38, which are associated with poor survival | 203 | 2.0 | <0.001 | 0.004 |
| FAELT_B_CLL_WITH_VH3_21_UP | Genes upregulated in samples from B-CLL with the immunoglobulin heavy chain VH3-21 gene | 39 | 1.71 | 0.009 | 0.038 |
| MOREAUX_MULTIPLE_MYELOMA _BY_TACI_UP | Up-regulated genes distinguishing in multiple myeloma samples with higher expression of TACI | 230 | 1.69 | 0.009 | 0.041 |
| TARTE_PLASMA_CELL_VS_PLASM ABLAST_UP | Genes upregulated in mature plasma cells compared with plasmablastic B lymphocytes | 255 | 1.73 | <0.001 | 0.031 |
| TARTE_PLASMA_CELL_VS_B_LYM PHOCYTE_UP | Genes upregulated in plasma cells compared with B lymphocytes | 74 | 1.68 | <0.001 | 0.043 |
| B. Not enriched in GPA | |||||
| SA_B_CELL_RECEPTOR_COMPLEX ES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases | 22 | 1.26 | 0.168 | 0.257 |
| ST_B_CELL_ANTIGEN_RECEPTOR | B Cell Antigen Receptor | 37 | 1.03 | 0.406 | 0.557 |
| MOREAUX_B_LYMPHOCYTE_MAT URATION_BY_TACI_UP | Genes upregulated in normal bone marrow plasma cells compared to polyclonal plasmablasts that also distinguished multiple myeloma samples by expression of levels of TACI | 50 | 0.86 | 0.710 | 0.813 |
| C. Enriched in healthy controls | |||||
| HUTTMANN_B_CLL_POOR_SURV IVAL_DN | Down-regulated genes in B-CLL patients expressing high levels of ZAP70 and CD38, which are associated with poor survival | 49 | 2.25 | <0.001 | 0.010 |
| FAELT_B_CLL_WITH_VH_REARRA NGEMENTS_DN | Genes downregulated in B-CLL patients with mutated immunoglobulin variable heavy chain genes | 44 | 1.76 | 0.002 | 0.033 |
GSEA showed that the genes involved in B cell activation and survival were enriched in patents with GPA as compared to healthy controls. B-CLL: B-cell chronic lymphocytic leukemia, FDR: false discovery rate, GPA: granulomatosis with polyangiitis, GSEA: gene set enrichment analysis, NES: normalized enrichment score, MAP: mitogen-activated protein, TACI: transmembrane activator and calcium modulator and cyclophylin ligand interactor.
* Description of gene sets are available on http://www.broadinstitute.org/gsea.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download