Abstract
Rheumatoid arthritis (RA) is a systemic inflammatory disease associated with both genetic and environmental factors. The DRB1 gene at the human leukocyte antigen (HLA) locus of chromosome 6p21.3 was the first genetic factor associated with RA to be identified in the 1980s; however, identification of causative genes other than those at the HLA locus has been challenging for geneticists because of the strong linkage disequilibrium in this locus and the non-Mendelian inheritance pattern of RA. Recent advances in high-throughput single nucleotide polymorphism genotyping technologies and bioinformatic analysis tools have facilitated the identification of positive associations of hundreds of genes with RA using family-based linkage analyses and genome wide association studies. Some of the RA associated genes at non-HLA loci are as follows: PADI4, PTPN22, STAT4, and TNFAIP3. In this paper, we describe the pathological mechanisms mediated by these genes. In addition, we review results of previous genetic studies of RA and future challenges in connecting the dots of missing heritability in the post-genomewide association study era.
Go to : 
REFERENCES
1. Dieudé P, Cornélis F. Genetic basis of rheumatoid arthritis. Joint Bone Spine. 2005; 72:520–6.
2. Korczowska I. Rheumatoid arthritis susceptibility genes: an overview. World J Orthop. 2014; 5:544–9.

3. Youinou P, Pers JO, Gershwin ME, Shoenfeld Y. Geo-epidemiology and autoimmunity. J Autoimmun. 2010; 34:J163–7.

4. Zvaifler NJ. The immunopathology of joint inflammation in rheumatoid arthritis. Adv Immunol. 1973; 16:265–336.

5. Schellekens GA, de Jong BA, van den Hoogen FH, van de Putte LB, van Venrooij WJ. Citrulline is an essential constituent of antigenic determinants recognized by rheumatoid arthritis-specific autoantibodies. J Clin Invest. 1998; 101:273–81.

6. MacGregor AJ, Snieder H, Rigby AS, Koskenvuo M, Kaprio J, Aho K, et al. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. 2000; 43:30–7.

7. Astorga GP, Williams RC Jr. Altered reactivity in mixed lymphocyte culture of lymphocytes from patients with rheumatoid arthritis. Arthritis Rheum. 1969; 12:547–54.

8. Stastny P. Association of the B-cell alloantigen DRw4 with rheumatoid arthritis. N Engl J Med. 1978; 298:869–71.

9. Legrand L, Lathrop GM, Marcelli-Barge A, Dryll A, Bardin T, Debeyre N, et al. HLA-DR genotype risks in seropositive rheumatoid arthritis. Am J Hum Genet. 1984; 36:690–9.
10. Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987; 30:1205–13.

11. Raychaudhuri S, Sandor C, Stahl EA, Freudenberg J, Lee HS, Jia X, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012; 44:291–6.

12. Viatte S, Plant D, Han B, Fu B, Yarwood A, Thomson W, et al. Association of HLA-DRB1 haplotypes with rheumatoid arthritis severity, mortality, and treatment response. JAMA. 2015; 313:1645–56.

13. McDaniel DO, Alarcón GS, Pratt PW, Reveille JD. Most African-American patients with rheumatoid arthritis do not have the rheumatoid antigenic determinant (epitope). Ann Intern Med. 1995; 123:181–7.

14. Teller K, Budhai L, Zhang M, Haramati N, Keiser HD, Davidson A. HLA-DRB1 and DQB typing of Hispanic American patients with rheumatoid arthritis: the “shared epitope” hypothesis may not apply. J Rheumatol. 1996; 23:1363–8.
15. Lee HS, Lee KW, Song GG, Kim HA, Kim SY, Bae SC. Increased susceptibility to rheumatoid arthritis in Koreans heterozygous for HLA-DRB1*0405 and *0901. Arthritis Rheum. 2004; 50:3468–75.

16. Wakitani S, Murata N, Toda Y, Ogawa R, Kaneshige T, Nishimura Y, et al. The relationship between HLA-DRB1 alleles and disease subsets of rheumatoid arthritis in Japanese. Br J Rheumatol. 1997; 36:630–6.

17. Kochi Y, Yamada R, Kobayashi K, Takahashi A, Suzuki A, Sekine A, et al. Analysis of single-nucleotide polymorphisms in Japanese rheumatoid arthritis patients shows additional susceptibility markers besides the classic shared epitope susceptibility sequences. Arthritis Rheum. 2004; 50:63–71.

18. Okamoto K, Makino S, Yoshikawa Y, Takaki A, Nagatsuka Y, Ota M, et al. Identification of I kappa BL as the second major histocompatibility complex-linked susceptibility locus for rheumatoid arthritis. Am J Hum Genet. 2003; 72:303–12.
19. Grennan DM, Dyer PA, Clague R, Dodds W, Smeaton I, Harris R. Family studies in RA – the importance of HLA-DR4 and of genes for autoimmune thyroid disease. J Rheumatol. 1983; 10:584–9.
20. Cornélis F, Fauré S, Martinez M, Prud'homme JF, Fritz P, Dib C, et al. ECRAF. New susceptibility locus for rheumatoid arthritis suggested by a genomewide linkage study. Proc Natl Acad Sci U S A. 1998; 95:10746–50.
21. Rossen RD, Brewer EJ, Sharp RM, Yunis EJ, Schanfield MS, Birdsall HH, et al. Familial rheumatoid arthritis: a kindred identified through a proband with seronegative juvenile arthritis includes members with seropositive, adult-onset disease. Hum Immunol. 1982; 4:183–96.

22. McDermott M, Molloy M, Cashin P, McMahon M, Spencer S, Jennings S, et al. A multicase family study of rheumatoid arthritis in south west Ireland. Dis Markers. 1986; 4:103–11.
23. Shaw MA, Clayton D, Atkinson SE, Williams H, Miller N, Sibthorpe D, et al. Linkage of rheumatoid arthritis to the candidate gene NRAMP1 on 2q35. J Med Genet. 1996; 33:672–7.

24. Jawaheer D, Seldin MF, Amos CI, Chen WV, Shigeta R, Monteiro J, et al. A genomewide screen in multiplex rheumatoid arthritis families suggests genetic overlap with other autoimmune diseases. Am J Hum Genet. 2001; 68:927–36.

25. Fisher SA, Lanchbury JS, Lewis CM. Meta-analysis of four rheumatoid arthritis genomewide linkage studies: con-firmation of a susceptibility locus on chromosome 16. Arthritis Rheum. 2003; 48:1200–6.

26. Amos CI, Chen WV, Lee A, Li W, Kern M, Lundsten R, et al. High-density SNP analysis of 642 Caucasian families with rheumatoid arthritis identifies two new linkage regions on 11p12 and 2q33. Genes Immun. 2006; 7:277–86.

27. Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003; 34:395–402.

28. Kochi Y, Thabet MM, Suzuki A, Okada Y, Daha NA, Toes RE, et al. PADI4 polymorphism predisposes male smokers to rheumatoid arthritis. Ann Rheum Dis. 2011; 70:512–5.

29. Remmers EF, Plenge RM, Lee AT, Graham RR, Hom G, Behrens TW, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007; 357:977–86.
30. Lee HS, Remmers EF, Le JM, Kastner DL, Bae SC, Gregersen PK. Association of STAT4 with rheumatoid arthritis in the Korean population. Mol Med. 2007; 13:455–60.

31. Kobayashi S, Ikari K, Kaneko H, Kochi Y, Yamamoto K, Shimane K, et al. Association of STAT4 with susceptibility to rheumatoid arthritis and systemic lupus erythematosus in the Japanese population. Arthritis Rheum. 2008; 58:1940–6.
32. Orozco G, Alizadeh BZ, Delgado-Vega AM, González-Gay MA, Balsa A, Pascual-Salcedo D, et al. Association of STAT4 with rheumatoid arthritis: a replication study in three European populations. Arthritis Rheum. 2008; 58:1974–80.
33. Bettelli E, Oukka M, Kuchroo VK. T(H)-17 cells in the circle of immunity and autoimmunity. Nat Immunol. 2007; 8:345–50.

34. Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004; 75:330–7.
35. Dai X, James RG, Habib T, Singh S, Jackson S, Khim S, et al. A disease-associated PTPN22 variant promotes systemic autoimmunity in murine models. J Clin Invest. 2013; 123:2024–36.

36. Zheng J, Ibrahim S, Petersen F, Yu X. Meta-analysis reveals an association of PTPN22 C1858T with autoimmune diseases, which depends on the localization of the affected tissue. Genes Immun. 2012; 13:641–52.

37. Thomson W, Barton A, Ke X, Eyre S, Hinks A, Bowes J, et al. Rheumatoid arthritis association at 6q23. Nat Genet. 2007; 39:1431–3.

38. Matmati M, Jacques P, Maelfait J, Verheugen E, Kool M, Sze M, et al. A20 (TNFAIP3) deficiency in myeloid cells triggers erosive polyarthritis resembling rheumatoid arthritis. Nat Genet. 2011; 43:908–12.

39. Okada Y, Terao C, Ikari K, Kochi Y, Ohmura K, Suzuki A, et al. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat Genet. 2012; 44:511–6.

40. Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, et al. Genomewide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010; 42:508–14.
41. Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014; 506:376–81.
Go to : 
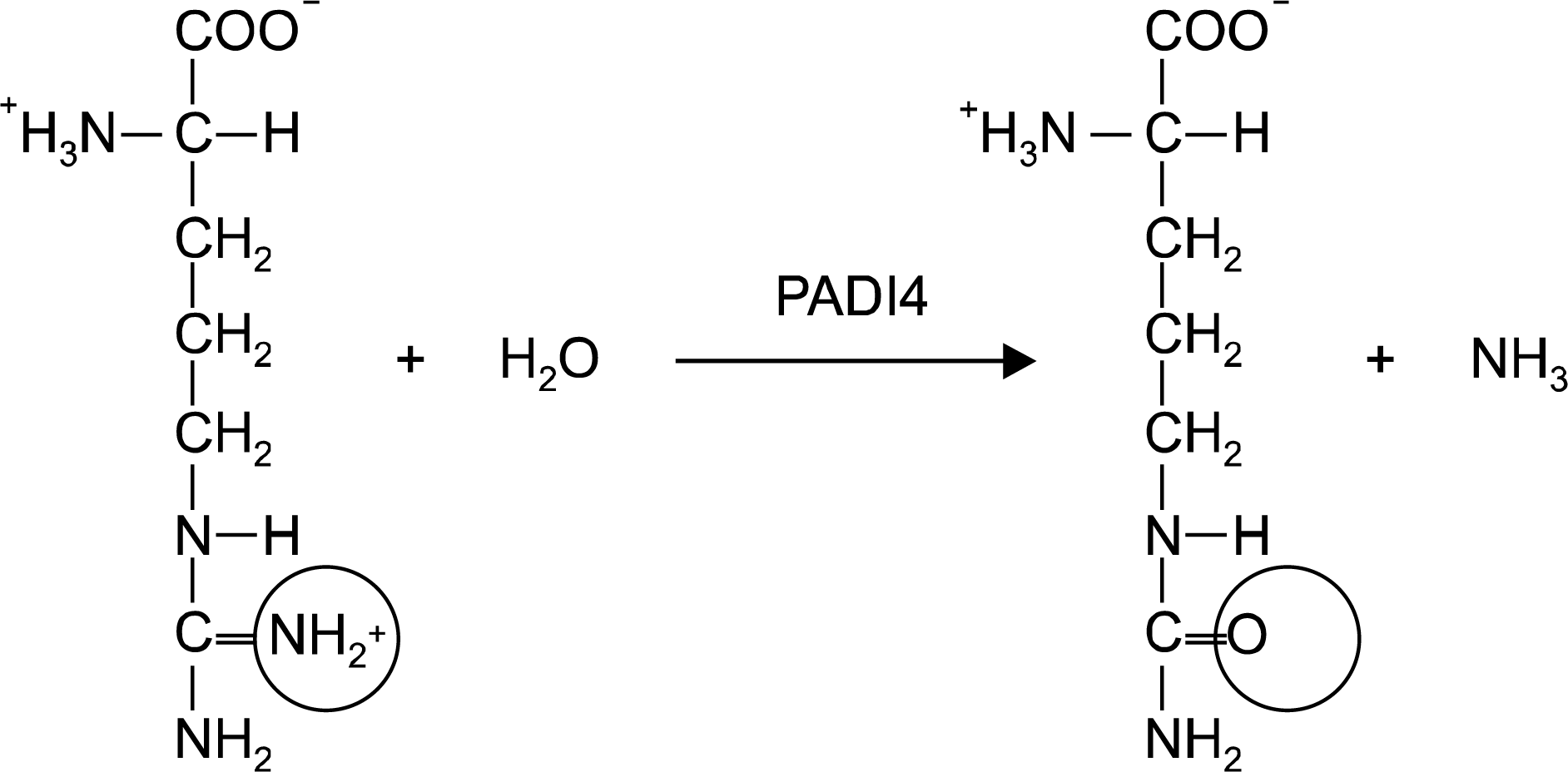 | Figure 1.Conversion of arginine residue to citrulline by peptidyl arginine deiminase, type IV (PADI4). |
Table 1.
Notable genetic studies of rheumatoid arthritis
GWAS: genomewide association study, HLA: human leukocyte antigen, MHC: major histocompatibility complex, RA: rheumatoid arthritis, SE: shared epitope, SNP: single nucleotide polymorphism, SLE: systemic lupus erythematosus, STAT4: signal transducer and activator of transcription 4, TNFAIP3: tumor necrosis factor-α-induced protein 3.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download