Abstract
Objective
We have previously shown that DICAM inhibits LPS-mediated macrophage differentiation. However, less is known about the exact action mechanisms of DICAM on the macrophage function and differentiation.
Methods
To induce differentiation into a resting M0 macrophage, THP-1 cells were cultured with 100 nM PMA for 24 h, and then rested for 3 days. THP-1 cells were infected with 50 moi of control LacZ- or DICAM-containing adenovirus. The RNA expression profile associated with DICAM during THP-1 differentiation was analyzed with a microarray chip and in silico analysis with Ingenuity Pathway Analysis (IPA) program.
Results
A disease and function analysis of the microarray data in DICAM-overexpressed THP-1 cells revealed a suppression in the expression of multiple genes involved in the response of myeloid cells and phagocytes, and an increase of genes associated with apoptosis of fibroblast cell-line, and viral infection and replication. The canonical pathway analysis also showed the most prominent changes of signaling pathways that involve inflammation responses. An upstream regulator analysis identifyingmolecules upstream of the genes that potentially explain the observed expression changes revealed that IRF7 and the genes in type 1 interferon system, such as IFNA2 and IFNAR, was significantly attenuated by DICAM. A mechanistic network analysis confirmed a direct causal association between IRF7 and type 1 interferon system. A real-time RT-PCR analysis validating the microarray data verified the significant suppression of IRFs, IFNA2, and IFNB1.
Go to : 
REFERENCES
1. Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013; 496:445–55.

2. Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010; 32:593–604.

3. Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008; 8:958–69.

4. Sica A, Schioppa T, Mantovani A, Allavena P. Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: potential targets of anticancer therapy. Eur J Cancer. 2006; 42:717–27.

6. Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med. 1992; 176:287–92.

7. Munder M, Eichmann K, Modolell M. Alternative metabolic states in murine macrophages reflected by the nitric oxide synthase/arginase balance: competitive regulation by CD4+ T cells correlates with Th1/Th2 phenotype. J Immunol. 1998; 160:5347–54.
8. de Weerd NA, Nguyen T. The interferons and their receptors–distribution and regulation. Immunol Cell Biol. 2012; 90:483–91.

9. Hu X, Chakravarty SD, Ivashkiv LB. Regulation of interferon and Toll-like receptor signaling during macrophage activation by opposing feedforward and feedback inhibition mechanisms. Immunol Rev. 2008; 226:41–56.

10. Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol. 2011; 11:750–61.

11. Hu X, Herrero C, Li WP, Antoniv TT, Falck-Pedersen E, Koch AE, et al. Sensitization of IFN-gamma Jak-STAT signaling during macrophage activation. Nat Immunol. 2002; 3:859–66.
12. Toshchakov V, Jones BW, Perera PY, Thomas K, Cody MJ, Zhang S, et al. TLR4, but not TLR2, mediates IFN-beta-induced STAT1alpha/beta-dependent gene expression in macrophages. Nat Immunol. 2002; 3:392–8.
13. Takaoka A, Mitani Y, Suemori H, Sato M, Yokochi T, Noguchi S, et al. Cross talk between interferon-gamma and -alpha/beta signaling components in caveolar membrane domains. Science. 2000; 288:2357–60.
14. Jung YK, Jeong JH, Ryoo HM, Kim HN, Kim YJ, Park EK, et al. Gene expression profile of human chondrocyte HCS-2/8 cell line by EST sequencing analysis. Gene. 2004; 330:85–92.

15. Jung YK, Jin JS, Jeong JH, Kim HN, Park NR, Choi JY. DICAM, a novel dual immunoglobulin domain containing cell adhesion molecule interacts with alphavbeta3 integrin. J Cell Physiol. 2008; 216:603–14.
16. Han SW, Jung YK, Lee EJ, Park HR, Kim GW, Jeong JH, et al. DICAM inhibits angiogenesis via suppression of AKT and p38 MAP kinase signalling. Cardiovasc Res. 2013; 98:73–82.

17. Jung YK, Han SW, Kim GW, Jeong JH, Kim HJ, Choi JY. DICAM inhibits osteoclast differentiation through attenuation of the integrin α Vβ 3 pathway. J Bone Miner Res. 2012; 27:2024–34.
18. Jung YK, Park HR, Lee EJ, Jeong DH, Kim GW, Choi JY, et al. DICAM Inhibits Activation of Macrophage by Lipopolysaccharide. J Rheum Dis. 2012; 19:196–205.

19. Stossi F, Madak-Erdoğan Z, Katzenellenbogen BS. Macrophage-elicited loss of estrogen receptor-α in breast cancer cells via involvement of MAPK and c-Jun at the ESR1 genomic locus. Oncogene. 2012; 31:1825–34.

20. Zhang L, Pagano JS. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol Cell Biol. 1997; 17:5748–57.

21. Schindler U, Hoey T, McKnight SL. Differentiation of T-helper lymphocytes: selective regulation by members of the STAT family of transcription factors. Genes Cells. 1996; 1:507–15.

22. Marié I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998; 17:6660–9.
23. Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, et al. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000; 13:539–48.
24. Génin P, Algarté M, Roof P, Lin R, Hiscott J. Regulation of RANTES chemokine gene expression requires coopera-tivity between NF-kappa B and IFN-regulatory factor transcription factors. J Immunol. 2000; 164:5352–61.
25. Lu R, Pitha PM. Monocyte differentiation to macrophage requires interferon regulatory factor 7. J Biol Chem. 2001; 276:45491–6.

26. Litvak V, Ratushny AV, Lampano AE, Schmitz F, Huang AC, Raman A, et al. A FOXO3-IRF7 gene regulatory cir-cuit limits inflammatory sequelae of antiviral responses. Nature. 2012; 490:421–5.

27. Weidinger C, Krause K, Mueller K, Klagge A, Fuhrer D. FOXO3 is inhibited by oncogenic PI3K/Akt signaling but can be reactivated by the NSAID sulindac sulfide. J Clin Endocrinol Metab. 2011; 96:E1361–71.

28. Yalcin S, Marinkovic D, Mungamuri SK, Zhang X, Tong W, Sellers R, et al. ROS-mediated amplification of AKT/mTOR signalling pathway leads to myeloproliferative syndrome in Foxo3(−/−) mice. EMBO J. 2010; 29:4118–31.

29. Tjiu JW, Chen JS, Shun CT, Lin SJ, Liao YH, Chu CY, et al. Tumor-associated macrophage-induced invasion and angiogenesis of human basal cell carcinoma cells by cy-clooxygenase-2 induction. J Invest Dermatol. 2009; 129:1016–25.

30. Zhang L, Dong Y, Dong Y, Cheng J, Du J. Role of in-tegrin-β 3 protein in macrophage polarization and regeneration of injured muscle. J Biol Chem. 2012; 287:6177–86.
32. Tailor P, Tamura T, Kong HJ, Kubota T, Kubota M, Borghi P, et al. The feedback phase of type I interferon induction in dendritic cells requires interferon regulatory factor 8. Immunity. 2007; 27:228–39.

33. Rönnblom L, Alm GV, Eloranta ML. Type I interferon and lupus. Curr Opin Rheumatol. 2009; 21:471–7.

34. Banchereau J, Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006; 25:383–92.

35. Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A. 2003; 100:2610–5.

36. Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003; 197:711–23.

37. Mathian A, Weinberg A, Gallegos M, Banchereau J, Koutouzov S. IFN-alpha induces early lethal lupus in pre-autoimmune (New Zealand Black × New Zealand White) F1 but not in BALB/c mice. J Immunol. 2005; 174:2499–506.
Go to : 
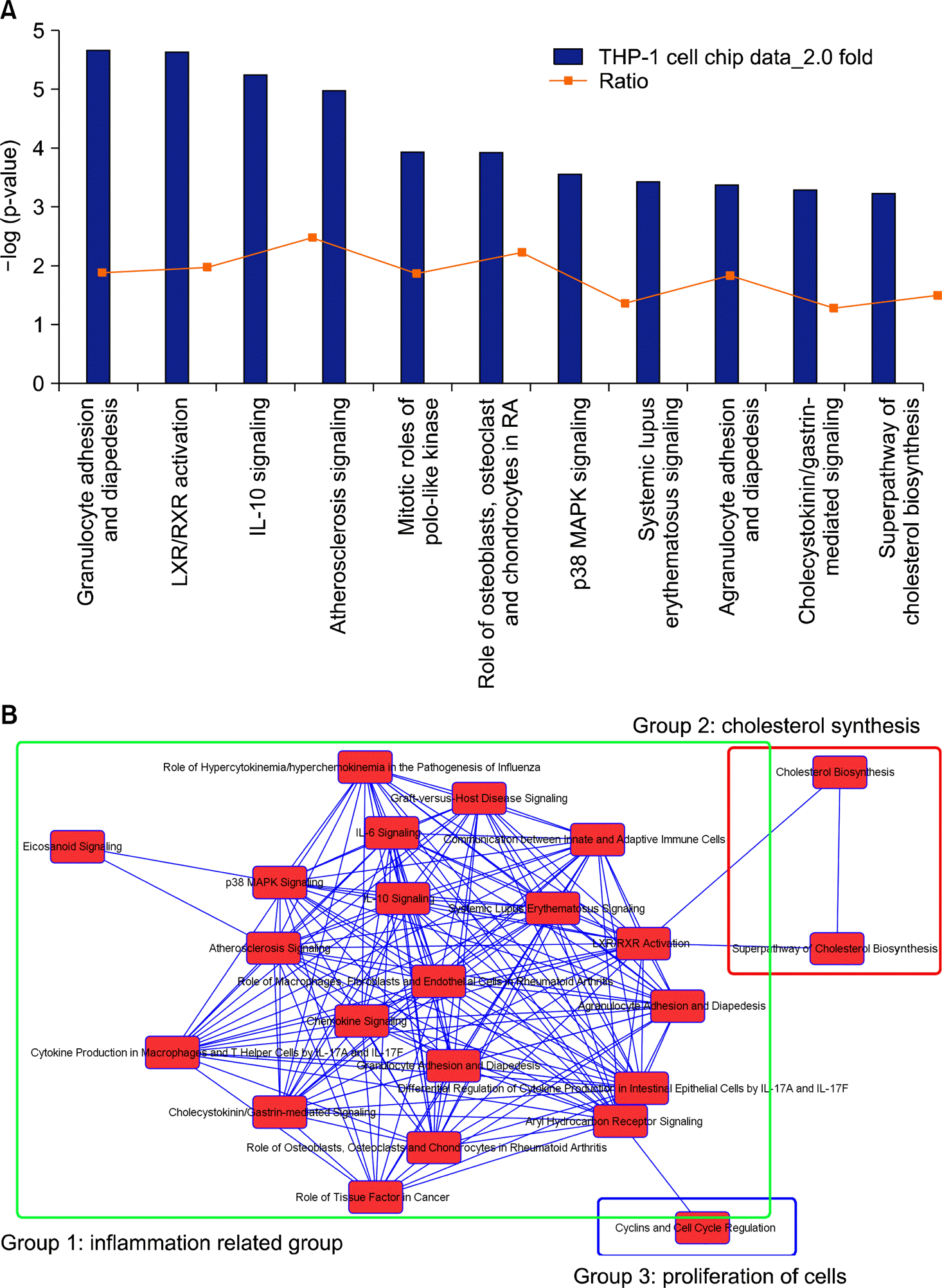 | Figure 1.Top hits for canonical pathways of differentially expressed proteins by DICAM and their overlap connections. (A) The most highly represented canonical pathways of genes differentially expressed during the macrophage differentiation of THP-1 cells. The columns represent the -log of the p-value calculated based on Fisher's exact test. The dot points represents the ratio of the number of genes in a given pathway that meet cut off criteria and total number of genes that make up that pathway. (B) The network of canonical pathways highlights the relationships between pathways and classify the functional domains. |
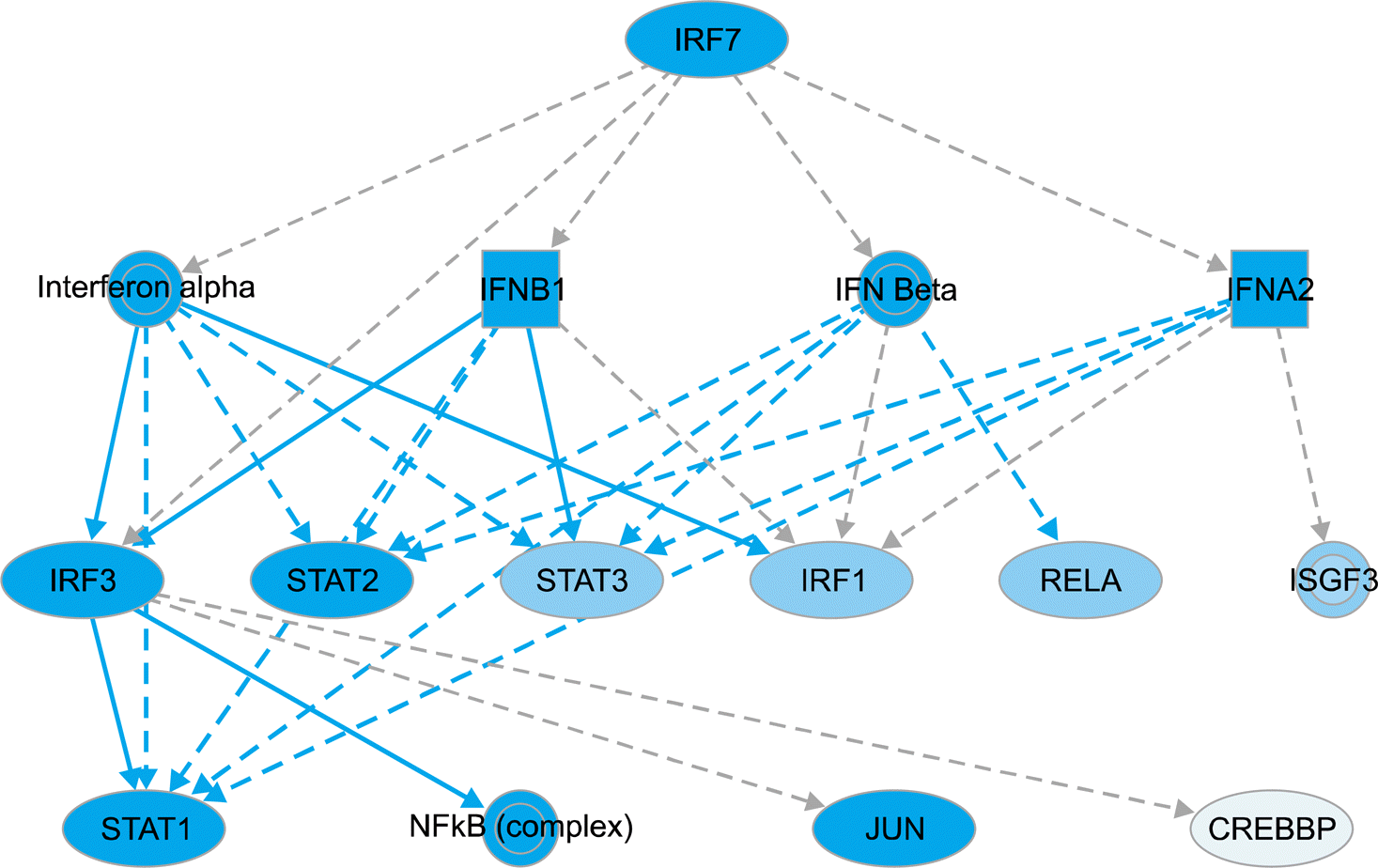 | Figure 2.IRF7-associated mechanistic networks of upstream regulators. IRF7, mostly suppressed by DICAM overexpression, is postu-lated to affect the expression of type I interferon and their downstream signaling molecules such as IRFs, STAT1/2/3 and RelA (p65). The set of total 15 regulators in mechanistic network has a connection with 187 dataset genes (not shown) among which 40 genes directly connect to IRF7. The blue color indicates a more confident predicted inhibition and the pale blue indicates a less confident predicted inhibition. |
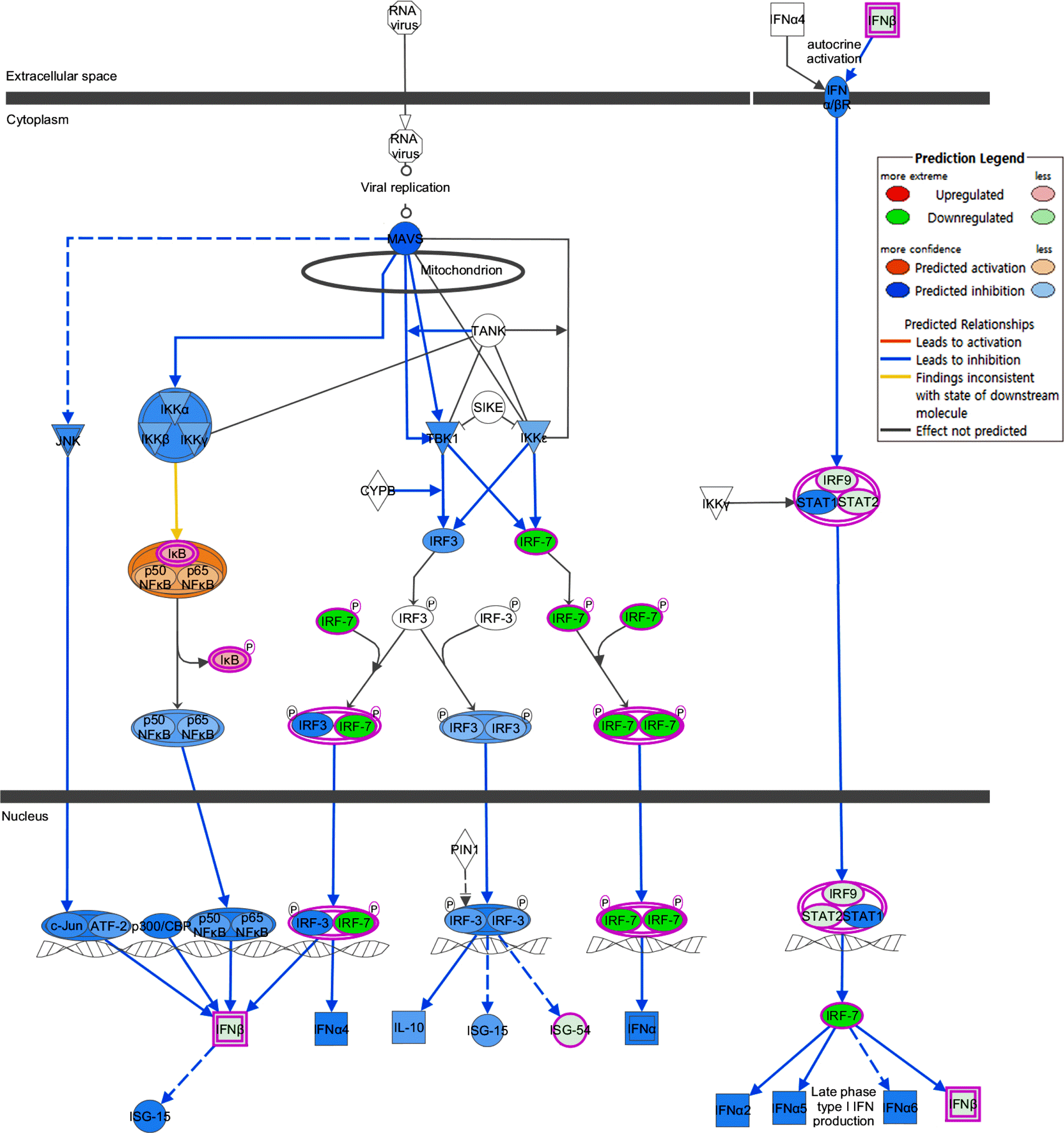 | Figure 3.The predicted DICAM-mediated inhibition of IRF canonical pathway. The canonical pathway of IRF activation by cytosolic pattern recognition receptor is analyzed in silico using Molecule Activity Predictor analysis of IPA. |
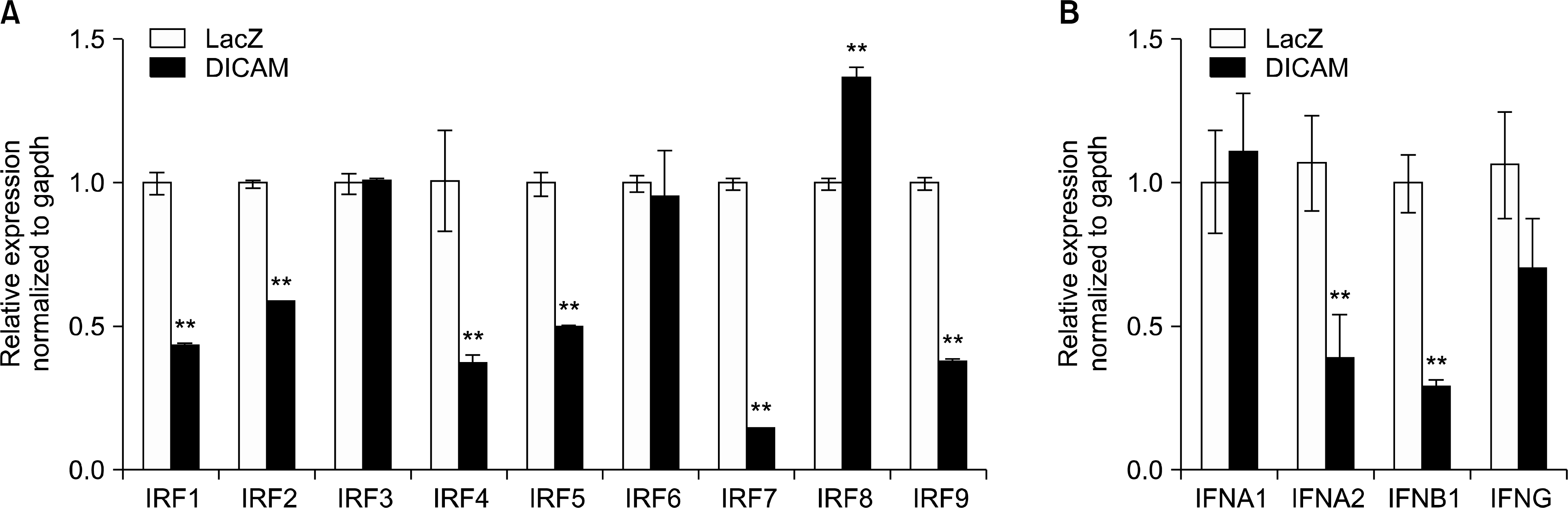 | Figure 4.DICAM-mediated inhibition of IRFs and type I interferon system. The microarray data is validated with real-time RT-PCR analysis. In the presence of PMA 100 nM, THP-1 cells infected with adenovirus encoding DICAM or LacZ for 24 h, and then left unstimulated for 3 days. Real-time RT-PCR analysis of (A) IRFs mRNA and (B) type I interferon (IFNA1, IFNA2 and IFNB1) and type 2 interferon (IFNG). |
Table 1.
The 8 mostly affected category of disease and biologic functions by DICAM overexpression in THP-1 cells that was analyzed with Downstream Effects Analysis using IPA
| Category | Disease or functions annotation | p-value* | Predicted activation state | Activation z-score† | #Molecules‡ |
|---|---|---|---|---|---|
| Cell-To-Cell Signaling and Interaction | Response of myeloid cells | 2.84E-05 | Decreased | −3.049 | 25 |
| Inflammatory Response | Response of phagocytes | 6.18E-06 | Decreased | −2.990 | 28 |
| Cell-To-Cell Signaling and Interaction | Response of phagocytes | 6.18E-06 | Decreased | −2.990 | 28 |
| Lipid Metabolism | Biosynthesis of polyunsaturated fatty acids | 1.30E-08 | Decreased | −2.951 | 35 |
| Cell Death and Survival | Apoptosis of fibroblast cell lines | 7.84E-06 | Increased | 3.126 | 40 |
| Infectious Disease | Viral Infection | 1.12E-09 | Increased | 3.095 | 168 |
| Infectious Disease | Replication of RNA virus | 1.33E-07 | Increased | 3.056 | 62 |
| Infectious Disease | Replication of virus | 3.98E-08 | Increased | 2.769 | 68 |
* The p-value was calculated with the Fischer's exact test, and reflects the likelyhood that the association between a set of genes in dataset and a related biological function is significant.
Table 2.
The 12 mostly activated and inhibited upstream regulators by DICAM overexpression in THP-1 cells predicted by the Upstream Regulator Analysis in IPA
| Upstream regulator | Fold change | Molecule type | Predicted activation state | Activation z-score* | p-value of overlap† |
|---|---|---|---|---|---|
| IRF7 | −38.745 | Tanscription regulator | Inhibited | −5.247 | 2.70E-18 |
| IFNA2 | Cytokine | Inhibited | −4.914 | 1.18E-15 | |
| Ifnar | Group | Inhibited | −4.636 | 1.58E-18 | |
| Tretinoin | Chemical – endogenous mammalian | Inhibited | −4.599 | 2.01E-19 | |
| IFNL1 | Cytokine | Inhibited | −4.463 | 4.80E-16 | |
| Lipopolysaccharide | Chemical drug | Inhibited | −4.345 | 4.02E-45 | |
| MYC | 2.052 | Transcription regulator | Activated | 5.330 | 4.19E-14 |
| TRIM24 | Transcription regulator | Activated | 4.208 | 7.73E-15 | |
| E2F1 | Transcription regulator | Activated | 4.060 | 6.09E-13 | |
| INSIG1 | −6.816 | Other | Activated | 3.957 | 3.90E-13 |
| TBX2 | Transcription regulator | Activated | 3.900 | 4.44E-10 | |
| IL1RN | −5.747 | Cytokine | Activated | 3.816 | 1.99E-11 |
IRF7: interferon regulatory factor7, IFNA2: interferon alpha-2, Ifnar: interferon-α/β receptor, IFNL1: interferon lambda1, MYC: v-myc avian myelocytomatosis viral oncogene homolog, TRIM24: Tripartite Motif Containing 24, E2F1: E2F transcription factor 1, INSIG1: insulin induced gene 1, TBX2: T-box transcription factor2, IL1RN: interleukin-1 receptor antagonist.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download