Abstract
Phenotypic characteristics of complex diseases such as rheumatoid arthritis are a consequence of interactions of genetic and environmental factors. Biomolecules closely interact with other molecular components and form functional modules, resulting in significant biologic action capability. While traditional biochemical research focuses on a single disease using narrowly constrained data, systems biology aims to interpret large volumes of highly complex and multilevel data obtained from high-through-put technologies to under-stand how biological systems function as a whole. Such a systems approach to complex diseases, so-called network medicine, can shape our comprehensive understanding of disease mechanisms by identifying modules temporally and spatially perturbed in the context of health and diseases. Given the unmet needs for diagnosis, monitoring, and treatment in rheumatoid arthritis, systems biology is obviously an emerging powerful tool to gain insight into disease mechanisms, study comorbidities, analyze therapeutic drugs and their targets, and discover novel network-based biomarkers.
Go to : 
References
1. Jin W, Qin P, Lou H, Jin L, Xu S. A systematic characterization of genes underlying both complex and Mendelian diseases. Hum Mol Genet. 2012; 21:1611–24.

2. Chuang HY, Hofree M, Ideker T. A decade of systems biology. Annu Rev Cell Dev Biol. 2010; 26:721–44.

4. Rubbert-Roth A, Finckh A. Treatment options in patients with rheumatoid arthritis failing initial TNF inhibitor therapy: a critical review. Arthritis Res Ther. 2009; 11(Suppl 1):S1.

5. Tanaka Y. Next stage of RA treatment: is TNF inhibitor- free remission a possible treatment goal? Ann Rheum Dis. 2013; 72(Suppl 2):ii124–7.
6. Lipsky PE, van der Heijde DM, St Clair EW, Furst DE, Breedveld FC, Kalden JR, et al. Antitumor Necrosis Factor Trial in Rheumatoid Arthritis with Concomitant Therapy Study Group. Infliximab and methotrexate in the treatment of rheumatoid arthritis. Antitumor Necrosis Factor Trial in Rheumatoid Arthritis with Concomitant Therapy Study Group. N Engl J Med. 2000; 343:1594–602.
7. Weinblatt ME, Keystone EC, Furst DE, Moreland LW, Weisman MH, Birbara CA, et al. Adalimumab, a fully human antitumor necrosis factor alpha monoclonal antibody, for the treatment of rheumatoid arthritis in patients taking concomitant methotrexate: the ARMADA trial. Arthritis Rheum. 2003; 48:35–45.
8. Weinblatt ME, Kremer JM, Bankhurst AD, Bulpitt KJ, Fleischmann RM, Fox RI, et al. A trial of etanercept, a recombinant tumor necrosis factor receptor:Fc fusion protein, in patients with rheumatoid arthritis receiving methotrexate. N Engl J Med. 1999; 340:253–9.

9. Finckh A, Simard JF, Gabay C, Guerne PA. SCQM physicians. Evidence for differential acquired drug resist-ance to anti-tumour necrosis factor agents in rheumatoid arthritis. Ann Rheum Dis. 2006; 65:746–52.

10. Prince FH, Bykerk VP, Shadick NA, Lu B, Cui J, Frits M, et al. Sustained rheumatoid arthritis remission is uncommon in clinical practice. Arthritis Res Ther. 2012; 14:R68.

11. Klarenbeek NB, van der Kooij SM, Gü ler-Yüksel M, van Groenendael JH, Han KH, Kerstens PJ, et al. Discontinuing treatment in patients with rheumatoid arthritis in sustained clinical remission: exploratory analyses from the BeSt study. Ann Rheum Dis. 2011; 70:315–9.

12. Aguilar-Lozano L, Castillo-Ortiz JD, Vargas-Serafin C, Morales-Torres J, Sanchez-Ortiz A, Sandoval-Castro C, et al. Sustained clinical remission and rate of relapse after tocilizumab withdrawal in patients with rheumatoid arthritis. J Rheumatol. 2013; 40:1069–73.

13. Ku CS, Loy EY, Pawitan Y, Chia KS. The pursuit of ge-nome-wide association studies: where are we now? J Hum Genet. 2010; 55:195–206.

14. Ormond KE, Wheeler MT, Hudgins L, Klein TE, Butte AJ, Altman RB, et al. Challenges in the clinical application of whole-genome sequencing. Lancet. 2010; 375:1749–51.

15. Wang J, Zhang Y, Marian C, Ressom HW. Identification of aberrant pathways and network activities from high-throughput data. Brief Bioinform. 2012; 13:406–19.

16. Barabási AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011; 12:56–68.

17. Likić VA, McConville MJ, Lithgow T, Bacic A. Systems biology: the next frontier for bioinformatics. Adv Bioinformatics. 2010; 268925.

18. Vaughan C. The rise of systems biology. UCSF Magazine. 2004; 24.
19. Ideker T, Galitski T, Hood L. A new approach to decoding life: systems biology. Annu Rev Genomics Hum Genet. 2001; 2:343–72.

20. MacLellan WR, Wang Y, Lusis AJ. Systems-based approaches to cardiovascular disease. Nat Rev Cardiol. 2012; 9:172–84.

22. Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick's Online Mendelian Inheritance in Man (OMIM). Nucleic Acids Res. 2009; 37:D793–6.
23. Gursoy A, Keskin O, Nussinov R. Topological properties of protein interaction networks from a structural perspective. Biochem Soc Trans. 2008; 36:1398–403.

24. Jeong H, Mason SP, Barabási AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001; 411:41–2.

25. Zotenko E, Mestre J, O'Leary DP, Przytycka TM. Why do hubs in the yeast protein interaction network tend to be essential: reexamining the connection between the network topology and essentiality. PLoS Comput Biol. 2008; 4:e1000140.

26. Zhu X, Gerstein M, Snyder M. Getting connected: analysis and principles of biological networks. Genes Dev. 2007; 21:1010–24.

27. Koutsogiannouli E, Papavassiliou AG, Papanikolaou NA. Complexity in cancer biology: is systems biology the an-swer? Cancer Med. 2013; 2:164–77.

28. Bauer-Mehren A, Bundschus M, Rautschka M, Mayer MA, Sanz F, Furlong LI. Gene-disease network analysis reveals functional modules in mendelian, complex and environmental diseases. PLoS One. 2011; 6:e20284.

29. Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabási AL. The human disease network. Proc Natl Acad Sci U S A. 2007; 104:8685–90.

30. Suthram S, Dudley JT, Chiang AP, Chen R, Hastie TJ, Butte AJ. Network-based elucidation of human disease similarities reveals common functional modules enriched for pluripotent drug targets. PLoS Comput Biol. 2010; 6:e1000662.

31. Cho DY, Kim YA, Przytycka TM. Chapter 5: Network biology approach to complex diseases. PLoS Comput Biol. 2012; 8:e1002820.

32. Navlakha S, Kingsford C. The power of protein interaction networks for associating genes with diseases. Bioinformatics. 2010; 26:1057–63.

33. Oti M, Snel B, Huynen MA, Brunner HG. Predicting disease genes using protein-protein interactions. J Med Genet. 2006; 43:691–8.

34. Wu X, Jiang R, Zhang MQ, Li S. Network-based global inference of human disease genes. Mol Syst Biol. 2008; 4:189.

35. Köhler S, Bauer S, Horn D, Robinson PN. Walking the interactome for prioritization of candidate disease genes. Am J Hum Genet. 2008; 82:949–58.

36. Vanunu O, Magger O, Ruppin E, Shlomi T, Sharan R. Associating genes and protein complexes with disease via network propagation. PLoS Comput Biol. 2010; 6:e1000641.

37. Chiche L, Jourde-Chiche N, Pascual V, Chaussabel D. Current perspectives on systems immunology approaches to rheumatic diseases. Arthritis Rheum. 2013; 65:1407–17.
38. Sirota M, Butte AJ. The role of bioinformatics in studying rheumatic and autoimmune disorders. Nat Rev Rheumatol. 2011; 7:489–94.

39. Toonen EJ, Barrera P, Radstake TR, van Riel PL, Scheffer H, Franke B, et al. Gene expression profiling in rheumatoid arthritis: current concepts and future directions. Ann Rheum Dis. 2008; 67:1663–9.

40. Viatte S, Plant D, Raychaudhuri S. Genetics and epi- genetics of rheumatoid arthritis. Nat Rev Rheumatol. 2013; 9:141–53.
41. Nakaoka H, Cui T, Tajima A, Oka A, Mitsunaga S, Kashiwase K, et al. A systems genetics approach provides a bridge from discovered genetic variants to biological pathways in rheumatoid arthritis. PLoS One. 2011; 6:e25389.

42. Sakaguchi N, Takahashi T, Hata H, Nomura T, Tagami T, Yamazaki S, et al. Altered thymic T-cell selection due to a mutation of the ZAP-70 gene causes autoimmune arthritis in mice. Nature. 2003; 426:454–60.

43. You S, Cho CS, Lee I, Hood L, Hwang D, Kim WU. A systems approach to rheumatoid arthritis. PLoS One. 2012; 7:e51508.

44. Keystone E, Burmester GR, Furie R, Loveless JE, Emery P, Kremer J, et al. Improvement in patient-reported outcomes in a rituximab trial in patients with severe rheumatoid arthritis refractory to antitumor necrosis factor therapy. Arthritis Rheum. 2008; 59:785–93.

45. Yoon HJ, You S, Yoo SA, Kim NH, Kwon HM, Yoon CH, et al. NF-AT5 is a critical regulator of inflammatory arthritis. Arthritis Rheum. 2011; 63:1843–52.

46. Wu G, Zhu L, Dent JE, Nardini C. A comprehensive molecular interaction map for rheumatoid arthritis. PLoS One. 2010; 5:e10137.

47. Xing H, McDonagh PD, Bienkowska J, Cashorali T, Runge K, Miller RE, et al. Causal modeling using network ensemble simulations of genetic and gene expression data predicts genes involved in rheumatoid arthritis. PLoS Comput Biol. 2011; 7:e1001105.

49. Venkatesan K, Rual JF, Vazquez A, Stelzl U, Lemmens I, Hirozane-Kishikawa T, et al. An empirical framework for binary interactome mapping. Nat Methods. 2009; 6:83–90.

50. Stumpf MP, Thorne T, de Silva E, Stewart R, An HJ, Lappe M, et al. Estimating the size of the human interactome. Proc Natl Acad Sci U S A. 2008; 105:6959–64.

51. Altelaar AF, Munoz J, Heck AJ. Next-generation proteomics: towards an integrative view of proteome dynamics. Nat Rev Genet. 2013; 14:35–48.

52. International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004; 431:931–45.
53. Jensen ON. Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry. Curr Opin Chem Biol. 2004; 8:33–41.
54. McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med. 2011; 365:2205–19.

55. Bottini N, Firestein GS. Duality of fibroblast-like syno-viocytes in RA: passive responders and imprinted aggressors. Nat Rev Rheumatol. 2013; 9:24–33.

56. Frenkel-Morgenstern M, Lacroix V, Ezkurdia I, Levin Y, Gabashvili A, Prilusky J, et al. Chimeras taking shape: potential functions of proteins encoded by chimeric RNA transcripts. Genome Res. 2012; 22:1231–42.

57. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011; 146:353–8.

58. Baker M. Proteomics: The interaction map. Nature. 2012; 484:271–5.
59. Bensimon A, Heck AJ, Aebersold R. Mass spectrome-try-based proteomics and network biology. Annu Rev Biochem. 2012; 81:379–405.

60. Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, et al. Inflammation and Host Response to Injury, Large Scale Collaborative Research Program. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013; 110:3507–12.
Go to : 
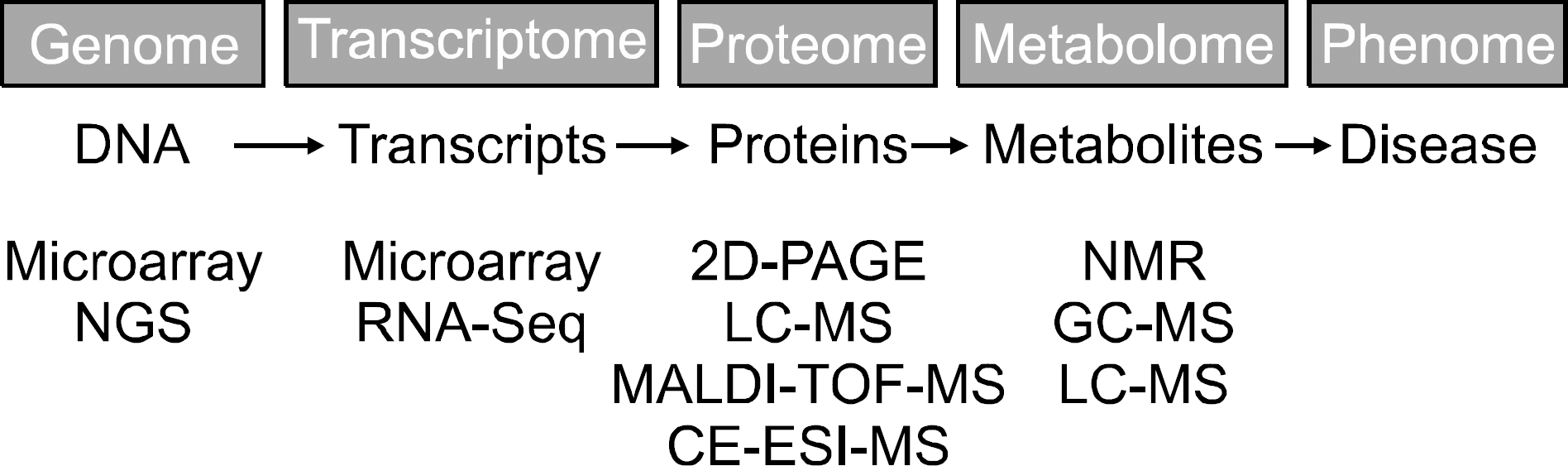 | Figure 1.Systems genetics and techniques for analysis of complex disease. NGS, next generation sequencing; 2D-PAGE, two-dimensional polyacrylamide gel electrophoresis; MALDI- TOF-MS, matrix-assisted laser desorption/ionization-time-of-fl ight mass spectrometry; CE-ESI-MS, capillary electrophoresis coupled with electrospray ionization mass spectrometry; NMR, nuclear magnetic resonance; GC-MS, gas chromatography-mass spectrometry; LC-MS, liquid chromatography-mass spectrometry. |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download