Abstract
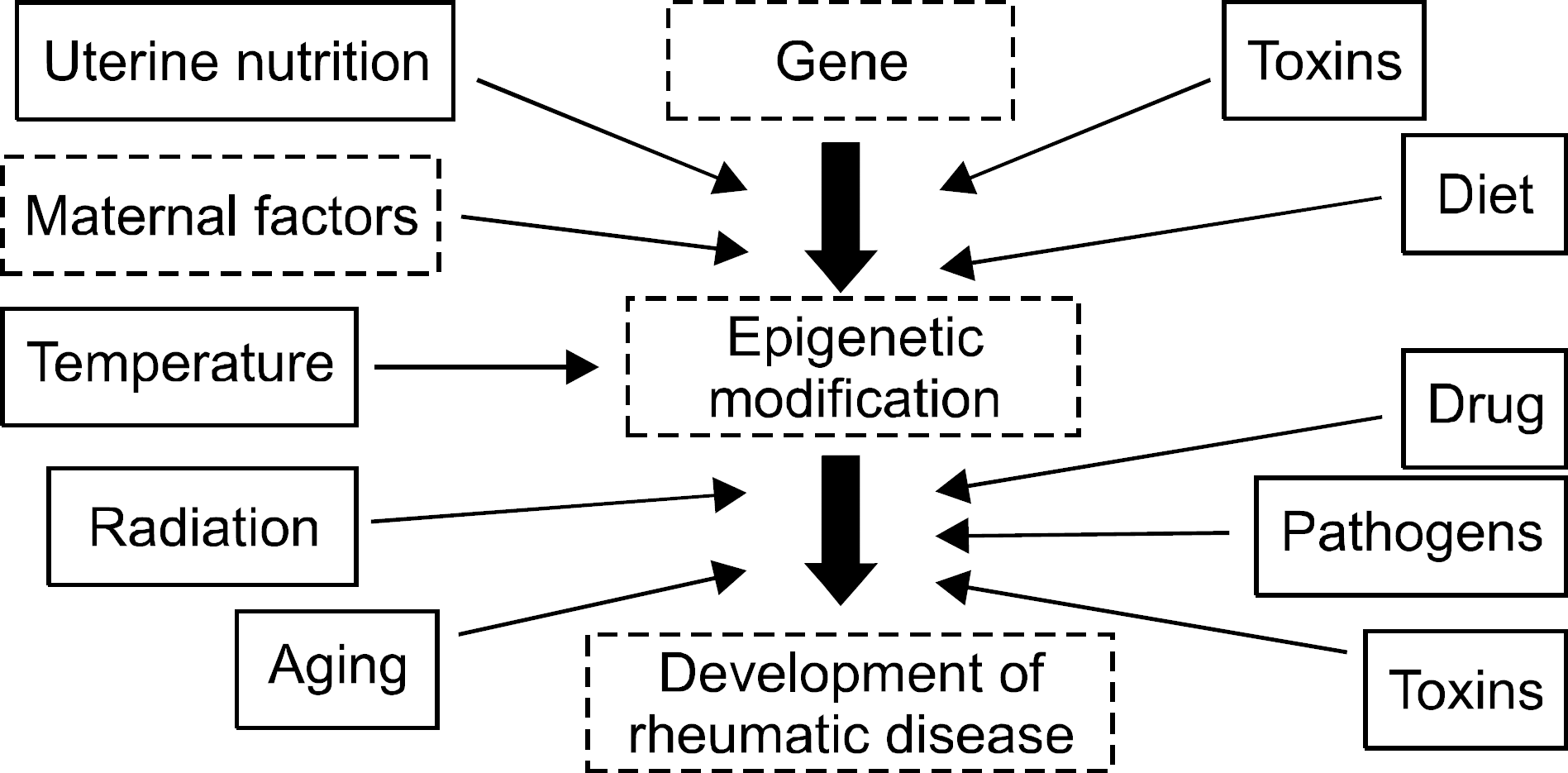
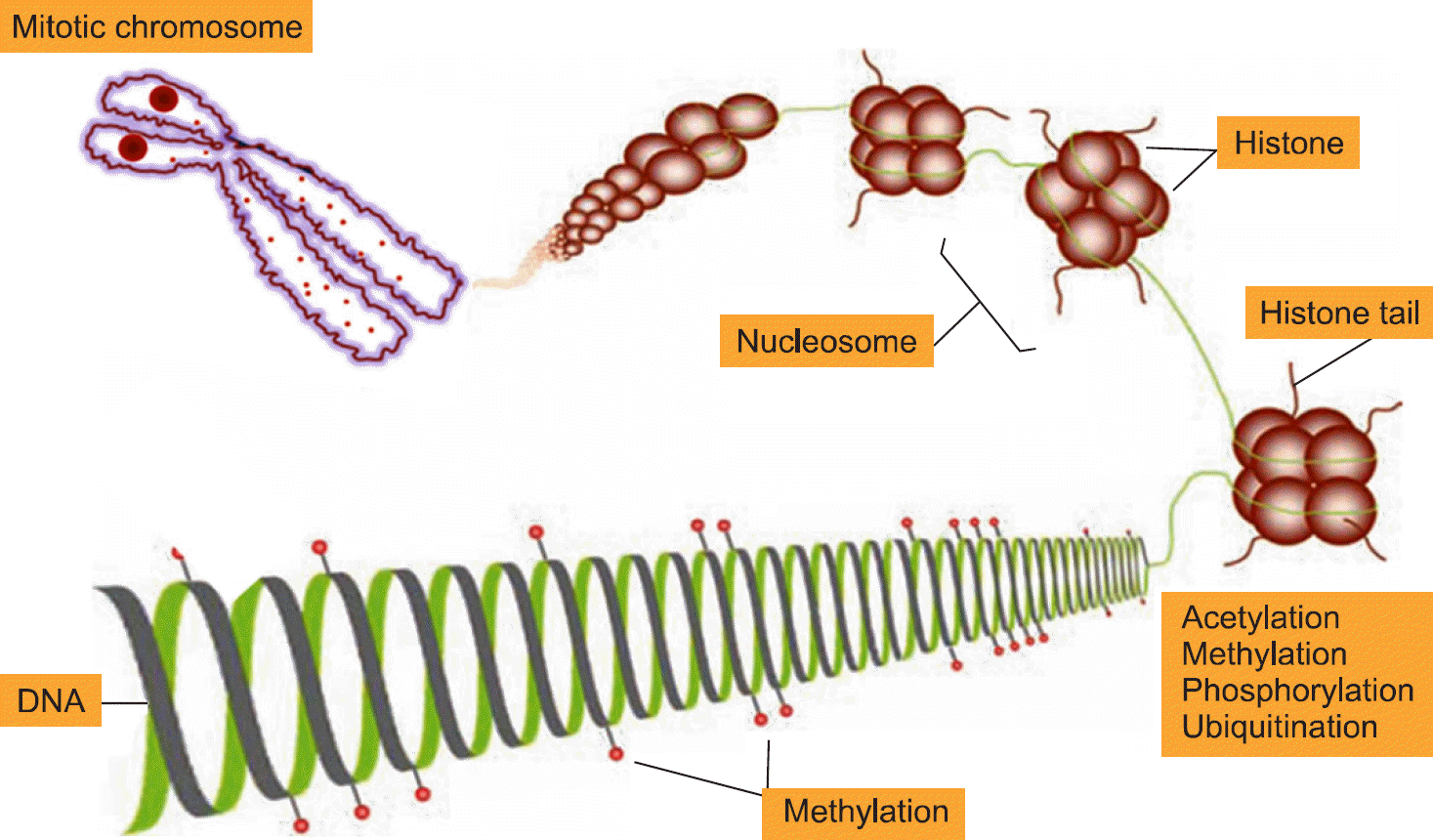
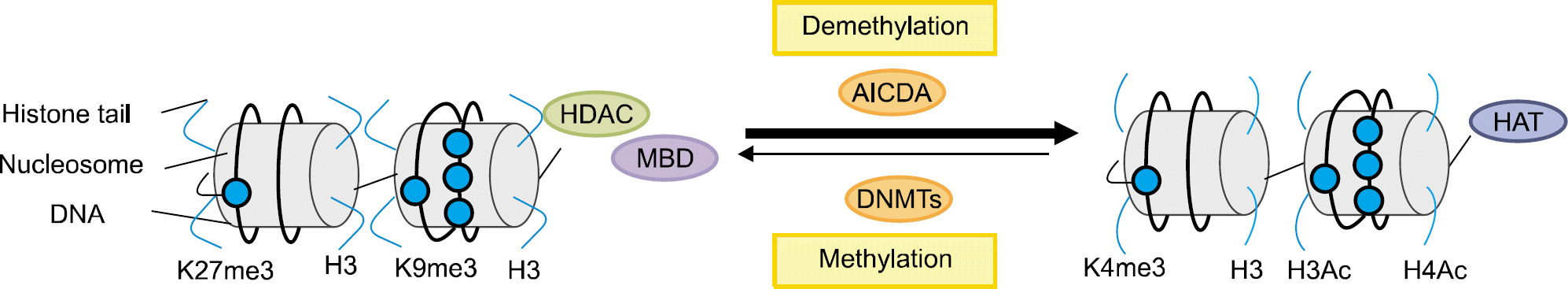
Epigenetics is defined as an inheritable effect that influen-ces gene activity, but does not involve a change in DNA sequence. Epigenetic gene regulation has an essential role in determining individual gene function and activity in each specific cell type. Epigenetics includes four predom-inant mechanisms: DNA methylation, histone modification, nucleosome positioning and microRNA (miRNA). These mechanisms influence gene expression, cell differentiation, proliferation, DNA repair and replication. Epigenetic modifications are far more sensitive to environmental stimuli than DNA sequence alterations. Candidate gene approaches have identified a small set of genes that undergo epigenetic changes, such as aberrant DNA demethylation, histone modification, as well as regulation by miRNA in rheumatic diseases. It is well known that T cells from patients with SLE or RA, as well as synovial fibroblasts from individuals with RA, have sequences undergoing DNA hypomethylation and/or histone modifications. In addition, miRNA regulates the gene expression by pairing with its target mRNAs and is often deregulated in systemic rheumatic diseases. High-throughput approaches are necessary for screening the epigenetic alterations, and it is essential to screen the specific tissue and cell types that are relevant to the disease pathogenesis. Identification of cell-specific targets of the epigenetic deregulation in rheumatic disorders will provide clinical markers for the diagnosis, disease progression and response to therapy. Our understanding of epigenetics is in its infancy. New generation of pharmaceuticals, which manipulate the epigenome to the switch targeted genes on or off are under investigation. The new field of repairing or optimizing the epigenome through epigenetic modifier and/or diet is wide open.
References
1. Quintero-Ronderos P, Montoya-Ortiz G. Epigenetics and autoimmune diseases. Autoimmune Dis. 2012; 2012; 593720.

2. Deapen D, Escalante A, Weinrib L, Horwitz D, Bachman B, Roy-Burman P, et al. A revised estimate of twin concordance in systemic lupus erythematosus. Arthritis Rheum. 1992; 35:311–8.
3. Järvinen P, Kaprio J, Mäkitalo R, Koskenvuo M, Aho K. Systemic lupus erythematosus and related systemic diseases in a nationwide twin cohort: an increased prevalence of disease in MZ twins and concordance of disease features. J Intern Med. 1992; 231:67–72.
4. Aho K, Koskenvuo M, Tuominen J, Kaprio J. Occurrence of rheumatoid arthritis in a nationwide series of twins. J Rheumatol. 1986; 13:899–902.
5. Bellamy N, Duffy D, Martin N, Mathews J. Rheumatoid arthritis in twins: a study of aetiopathogenesis based on the Australian Twin Registry. Ann Rheum Dis. 1992; 51:588–93.

6. Silman AJ, MacGregor AJ, Thomson W, Holligan S, Carthy D, Farhan A, et al. Twin concordance rates for rheumatoid arthritis: results from a nationwide study. Br J Rheumatol. 1993; 32:903–7.

7. Fan S, Zhang X. CpG island methylation pattern in different human tissues and its correlation with gene expression. Biochem Biophys Res Commun. 2009; 383:421–5.

10. Gregory PD, Wagner K, Hörz W. Histone acetylation and chromatin remodeling. Exp Cell Res. 2001; 265:195–202.

11. Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008; 132:887–98.

12. Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010; 466:388–92.

13. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–97.
14. Lu Q, Kaplan M, Ray D, Ray D, Zacharek S, Gutsch D, et al. Demethylation of ITGAL (CD11a) regulatory sequences in systemic lupus erythematosus. Arthritis Rheum. 2002; 46:1282–91.
15. Lu Q, Wu A, Tesmer L, Ray D, Yousif N, Richardson B. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J Immunol. 2007; 179:6352–8.

16. Kaplan MJ, Lu Q, Wu A, Attwood J, Richardson B. Demethylation of promoter regulatory elements contributes to perforin overexpression in CD4+ lupus T cells. J Immunol. 2004; 172:3652–61.

17. Oelke K, Lu Q, Richardson D, Wu A, Deng C, Hanash S, et al. Overexpression of CD70 and overstimulation of IgG synthesis by lupus T cells and T cells treated with DNA methylation inhibitors. Arthritis Rheum. 2004; 50:1850–60.

18. Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, et al. Changes in the pattern of DNA methylation associate with twin dis-cordance in systemic lupus erythematosus. Genome Res. 2010; 20:170–9.

19. Garaud S, Le Dantec C, Jousse-Joulin S, Hanrotel-Saliou C, Saraux A, Mageed RA, et al. IL-6 modulates CD5 expression in B cells from patients with lupus by regulating DNA methylation. J Immunol. 2009; 182:5623–32.

20. Neidhart M, Rethage J, Kuchen S, Künzler P, Crowl RM, Billingham ME, et al. Retrotransposable L1 elements expressed in rheumatoid arthritis synovial tissue: association with genomic DNA hypomethylation and influence on gene expression. Arthritis Rheum. 2000; 43:2634–47.

21. Nile CJ, Read RC, Akil M, Duff GW, Wilson AG. Methylation status of a single CpG site in the IL6 promoter is related to IL6 messenger RNA levels and rheumatoid arthritis. Arthritis Rheum. 2008; 58:2686–93.
22. Takami N, Osawa K, Miura Y, Komai K, Taniguchi M, Shiraishi M, et al. Hypermethylated promoter region of DR3, the death receptor 3 gene, in rheumatoid arthritis synovial cells. Arthritis Rheum. 2006; 54:779–87.
23. Kubo M, Czuwara-Ladykowska J, Moussa O, Markiewicz M, Smith E, Silver RM, et al. Persistent down-regulation of Fli1, a suppressor of collagen transcription, in fibrotic scleroderma skin. Am J Pathol. 2003; 163:571–81.

24. Dieker JW, Fransen JH, van Bavel CC, Briand JP, Jacobs CW, Muller S, et al. Apoptosis-induced acetylation of histones is pathogenic in systemic lupus erythematosus. Arthritis Rheum. 2007; 56:1921–33.

25. van Bavel CC, Dieker JW, Tamboer WP, van der Vlag J, Berden JH. Lupus-derived monoclonal autoantibodies against apoptotic chromatin recognize acetylated confor- mational epitopes. Mol Immunol. 2010; 48:248–56.
26. Hu N, Qiu X, Luo Y, Yuan J, Li Y, Lei W, et al. Abnormal histone modification patterns in lupus CD4+ T cells. J Rheumatol. 2008; 35:804–10.
27. Young DA, Lakey RL, Pennington CJ, Jones D, Kevor-kian L, Edwards DR, et al. Histone deacetylase inhibitors modulate metalloproteinase gene expression in chondrocytes and block cartilage resorption. Arthritis Res Ther. 2005; 7:R503–12.
28. Nishida K, Komiyama T, Miyazawa S, Shen ZN, Furumatsu T, Doi H, et al. Histone deacetylase inhibitor sup-pression of autoantibody-mediated arthritis in mice via regulation of p16INK4a and p21(WAF1/Cip1) expression. Arthritis Rheum. 2004; 50:3365–76.

29. Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, et al. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009; 60:1065–75.

30. Zhao X, Tang Y, Qu B, Cui H, Wang S, Wang L, et al. MicroRNA-125a contributes to elevated inflammatory chemokine RANTES levels via targeting KLF13 in systemic lupus erythematosus. Arthritis Rheum. 2010; 62:3425–35.

31. Pan W, Zhu S, Yuan M, Cui H, Wang L, Luo X, et al. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J Immunol. 2010; 184:6773–81.
32. Nakamachi Y, Kawano S, Takenokuchi M, Nishimura K, Sakai Y, Chin T, et al. MicroRNA-124a is a key regulator of proliferation and monocyte chemoattractant protein 1 secretion in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheum. 2009; 60:1294–304.

33. Kawano S, Nakamachi Y. miR-124a as a key regulator of proliferation and MCP-1 secretion in synoviocytes from patients with rheumatoid arthritis. Ann Rheum Dis. 2011; 70(Suppl 1):i88–91.

34. Stanczyk J, Ospelt C, Karouzakis E, Filer A, Raza K, Kolling C, et al. Altered expression of microRNA-203 in rheumatoid arthritis synovial fibroblasts and its role in fi-broblast activation. Arthritis Rheum. 2011; 63:373–81.

35. Stanczyk J, Pedrioli DM, Brentano F, Sanchez-Pernaute O, Kolling C, Gay RE, et al. Altered expression of Micro-RNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008; 58:1001–9.

36. Alevizos I, Illei GG. MicroRNAs in Sjögren's syndrome as a prototypic autoimmune disease. Autoimmun Rev. 2010; 9:618–21.

37. Barreto G, Schäfer A, Marhold J, Stach D, Swaminathan SK, Handa V, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007; 445:671–5.

38. Fritz EL, Papavasiliou FN. Cytidine deaminases: AIDing DNA demethylation? Genes Dev. 2010; 24:2107–14.

39. Lee BH, Yegnasubramanian S, Lin X, Nelson WG. Procainamide is a specific inhibitor of DNA methyltransferase 1. J Biol Chem. 2005; 280:40749–56.

40. Deng C, Lu Q, Zhang Z, Rao T, Attwood J, Yung R, et al. Hydralazine may induce autoimmunity by inhibiting extracellular signal-regulated kinase pathway signaling. Arthritis Rheum. 2003; 48:746–56.

41. Yung R, Chang S, Hemati N, Johnson K, Richardson B. Mechanisms of drug-induced lupus. IV. Comparison of procainamide and hydralazine with analogs in vitro and in vivo. Arthritis Rheum. 1997; 40:1436–43.

42. Fu LH, Ma CL, Cong B, Li SJ, Chen HY, Zhang JG. Hypomethylation of proximal CpG motif of interleukin-10 promoter regulates its expression in human rheumatoid arthritis. Acta Pharmacol Sin. 2011; 32:1373–80.

43. Bruns A, Bläss S, Hausdorf G, Burmester GR, Hiepe F. Nucleosomes are major T and B cell autoantigens in systemic lupus erythematosus. Arthritis Rheum. 2000; 43:2307–15.

44. Dai Y, Zhang L, Hu C, Zhang Y. Genome-wide analysis of histone H3 lysine 4 trimethylation by ChIP-chip in peripheral blood mononuclear cells of systemic lupus erythematosus patients. Clin Exp Rheumatol. 2010; 28:158–68.
45. Zhang Z, Song L, Maurer K, Petri MA, Sullivan KE. Global H4 acetylation analysis by ChIP-chip in systemic lupus erythematosus monocytes. Genes Immun. 2010; 11:124–33.

46. Manabe H, Nasu Y, Komiyama T, Furumatsu T, Kitamura A, Miyazawa S, et al. Inhibition of histone deacetylase down-regulates the expression of hypoxia-induced vascular endothelial growth factor by rheumatoid synovial fibroblasts. Inflamm Res. 2008; 57:4–10.

47. Nasu Y, Nishida K, Miyazawa S, Komiyama T, Kadota Y, Abe N, et al. Trichostatin A, a histone deacetylase inhibitor, suppresses synovial inflammation and subsequent cartilage destruction in a collagen antibody-induced arthritis mouse model. Osteoarthritis Cartilage. 2008; 16:723–32.

48. Grabiec AM, Korchynskyi O, Tak PP, Reedquist KA. Histone deacetylase inhibitors suppress rheumatoid arthritis fibroblast-like synoviocyte and macrophage IL-6 production by accelerating mRNA decay. Ann Rheum Dis. 2012; 71:424–31.

49. Angelov D, Molla A, Perche PY, Hans F, Côté J, Khochbin S, et al. The histone variant macroH2A inter-feres with transcription factor binding and SWI/SNF nucleosome remodeling. Mol Cell. 2003; 11:1033–41.

Table 1.
Concordance rate of autoimmune diseases between monozygotic twins
| Disease | Concordance rate | References |
|---|---|---|
| Systmeic lupus erythematosus | 11∼25% | (2, 3) |
| Rheumatoid arthritis | 12∼22% | (4–6) |
Table 2.
Epigenetic modification in autoimmune diseases
Table 3.
Hypomethylated genes in autoimmune diseases




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download