Abstract
Objective
The physiological and pathogenetic role of microRNAs (miRNAs) in the maintenance of joint homeostasis and in the development of arthritis is recently being elucidated. In this study, we attempted to identify differentially expressed miRNAs in human osteoarthritis (OA) chondrocytes in response to interleukin (IL)-1β. In addition, simultaneous profiling of miRNA and mRNA expression was performed to get an integrated analysis of miRNA and mRNA expression.
Methods
Monolayer cultured chondrocytes obtained from knee cartilages of OA patients were stimulated with IL-1β for 4 hours and RNA was isolated. One microgram of total RNA was polyadenylated and converted to cDNA and miRNA microarray was performed. Seven hundred thirty five oligos were used, corresponding to 470 well-annotated human miRNA sequences and 265 potential miRNAs that were identified recently. mRNA microarray was performed simultaneously using the RNA samples that were used for miRNA array. Both sequence and expression information was used to identify regulatory relationship between miRNA and mRNA pairs.
Results
Expression profiling of miRNA extracted from IL-1β treated chondrocytes identified 25 miRNA which showed differential expression. We also identified 7190 mRNAs differentially regulated by IL-1β treatment. Among the 25 miRNAs differentially regulated, 13 miRNAs had targets searched by MiRANDA scheme. By combining target search and miRNA-mRNA pairing, we could identify 1043 miRNA-mRNA target pairs. MiR-200a was found to be expressed in human OA and normal cartilages, with downregulation in OA lesion cartilages.
References
2. Daheshia M, Yao JQ. The interleukin 1beta pathway in the pathogenesis of osteoarthritis. J Rheumatol. 2008; 35:2306–12.
3. Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene. 2006; 25:6163–9.

4. Arora A, Simpson DA. Individual mRNA expression profiles reveal the effects of specific microRNAs. Genome Biol. 2008; 9:R82.

5. Sato F, Tsuchiya S, Meltzer SJ, Shimizu K. MicroRNAs and epigenetics. FEBS J. 2011; 278:1598–609.

6. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–97.
8. Huang JC, Babak T, Corson TW, Chua G, Khan S, Gallie BL, et al. Using expression profiling data to identify human microRNA targets. Nat Methods. 2007; 4:1045–9.

9. Kim HA, Cho ML, Choi HY, Yoon CS, Jhun JY, Oh HJ, et al. The catabolic pathway mediated by Toll-like receptors in human osteoarthritic chondrocytes. Arthritis Rheum. 2006; 54:2152–63.

10. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statisti Soc B. 1995; 57:289–300.

11. John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004; 2:e363.

12. Tallheden T, Karlsson C, Brunner A, Van Der Lee J, Hagg R, Tommasini R, et al. Gene expression during redifferentiation of human articular chondrocytes. Osteoarthritis Cartilage. 2004; 12:525–35.

13. Isomäki P, Alanärä T, Isohanni P, Lagerstedt A, Korpela M, Moilanen T, et al. The expression of SOCS is altered in rheumatoid arthritis. Rheumatology (Oxford). 2007; 46:1538–46.
14. Kan A, Ikeda T, Saito T, Yano F, Fukai A, Hojo H, et al. Screening of chondrogenic factors with a real-time flu-orescence-monitoring cell line ATDC5-C2ER: identification of sorting nexin 19 as a novel factor. Arthritis Rheum. 2009; 60:3314–23.

15. Badariotti F, Kypriotou M, Lelong C, Dubos MP, Renard E, Galera P, et al. The phylogenetically conserved mollus-can chitinase-like protein 1 (Cg-Clp1), homologue of human HC-gp39, stimulates proliferation and regulates synthesis of extracellular matrix components of mammalian chondrocytes. J Biol Chem. 2006; 281:29583–96.

16. Maneiro E, López-Armada MJ, de Andres MC, Caramés B, Martín MA, Bonilla A, et al. Effect of nitric oxide on mitochondrial respiratory activity of human articular chondrocytes. Ann Rheum Dis. 2005; 64:388–95.

17. Albrecht AN, Schwabe GC, Stricker S, Böddrich A, Wanker EE, Mundlos S. The synpolydactyly homolog (spdh) mutation in the mouse – a defect in patterning and growth of limb cartilage elements. Mech Dev. 2002; 112:53–67.

18. Sironen RK, Karjalainen HM, Tö rrönen K, Elo MA, Kaarniranta K, Takigawa M, et al. High pressure effects on cellular expression profile and mRNA stability. A cDNA array analysis. Biorheology. 2002; 39:111–7.
19. Karjalainen HM, Sironen RK, Elo MA, Kaarniranta K, Takigawa M, Helminen HJ, et al. Gene expression profiles in chondrosarcoma cells subjected to cyclic stretching and hydrostatic pressure. A cDNA array study. Biorheology. 2003; 40:93–100.
20. Mienaltowski MJ, Huang L, Stromberg AJ, MacLeod JN. Differential gene expression associated with postnatal equine articular cartilage maturation. BMC Musculoskelet Disord. 2008; 9:149.

21. Millward-Sadler SJ, Khan NS, Bracher MG, Wright MO, Salter DM. Roles for the interleukin-4 receptor and associated JAK/STAT proteins in human articular chondrocyte mechanotransduction. Osteoarthritis Cartilage. 2006; 14:991–1001.

22. Lee CR, Sakai D, Nakai T, Toyama K, Mochida J, Alini M, et al. A phenotypic comparison of intervertebral disc and articular cartilage cells in the rat. Eur Spine J. 2007; 16:2174–85.

23. Yamaoka H, Nishizawa S, Asawa Y, Fujihara Y, Ogasawara T, Yamaoka K, et al. Involvement of fibroblast growth factor 18 in dedifferentiation of cultured human chondrocytes. Cell Prolif. 2010; 43:67–76.

24. Moore EE, Bendele AM, Thompson DL, Littau A, Waggie KS, Reardon B, et al. Fibroblast growth factor-18 stimulates chondrogenesis and cartilage repair in a rat model of injury-induced osteoarthritis. Osteoarthritis Cartilage. 2005; 13:623–31.

25. Hiscock DR, Caterson B, Flannery CR. Expression of hyaluronan synthases in articular cartilage. Osteoarthritis Cartilage. 2000; 8:120–6.

26. Long D, Blake S, Song XY, Lark M, Loeser RF. Human articular chondrocytes produce IL-7 and respond to IL-7 with increased production of matrix metalloproteinase-13. Arthritis Res Ther. 2008; 10:R23.

27. Nifuji A, Miura N, Kato N, Kellermann O, Noda M. Bone morphogenetic protein regulation of fork-head/winged helix transcription factor Foxc2 (Mfh1) in a murine mesodermal cell line C1 and in skeletal precursor cells. J Bone Miner Res. 2001; 16:1765–71.

28. Bursell L, Woods A, James CG, Pala D, Leask A, Beier F. Src kinase inhibition promotes the chondrocyte phenotype. Arthritis Res Ther. 2007; 9:R105.

29. Wang Y, Toury R, Hauchecorne M, Balmain N. Expression and subcellular localization of the Myc super-family proteins: c-Myc, Max, Mad1 and Mxi1 in the epi-physeal plate cartilage chondrocytes of growing rats. Cell Mol Biol (Noisy-le-grand). 1997; 43:175–88.
30. Aguilar A, Wu S, De Luca F. P450 oxidoreductase expressed in rat chondrocytes modulates chondrogenesis via cholesterol- and Indian Hedgehog-dependent mechanisms. Endocrinology. 2009; 150:2732–9.

31. Lefebvre V, Smits P. Transcriptional control of chondrocyte fate and differentiation. Birth Defects Res C Embryo Today. 2005; 75:200–12.

32. Watanabe Y, Namba A, Honda K, Aida Y, Matsumura H, Shimizu O, et al. IL-1beta stimulates the expression of prostaglandin receptor EP4 in human chondrocytes by increasing production of prostaglandin E2. Connect Tissue Res. 2009; 50:186–93.
33. Li X, Ellman M, Muddasani P, Wang JH, Cs-Szabo G, van Wijnen AJ, et al. Prostaglandin E2 and its cognate EP receptors control human adult articular cartilage homeostasis and are linked to the pathophysiology of osteoarthritis. Arthritis Rheum. 2009; 60:513–23.
34. Otsuka S, Aoyama T, Furu M, Ito K, Jin Y, Nasu A, et al. PGE2 signal via EP2 receptors evoked by a selective agonist enhances regeneration of injured articular cartilage. Osteoarthritis Cartilage. 2009; 17:529–38.

35. Barre PE, Redini F, Boumediene K, Vielpeau C, Pujol JP. Semiquantitative reverse transcription-polymerase chain reaction analysis of syndecan-1 and −4 messages in cartilage and cultured chondrocytes from osteoarthritic joints. Osteoarthritis Cartilage. 2000; 8:34–43.

36. Echtermeyer F, Bertrand J, Dreier R, Meinecke I, Neugebauer K, Fuerst M, et al. Syndecan-4 regulates ADAMTS-5 activation and cartilage breakdown in osteoarthritis. Nat Med. 2009; 15:1072–6.

37. Zhang L, Yang M, Yang D, Cavey G, Davidson P, Gibson G. Molecular interactions of MMP-13 C-terminal domain with chondrocyte proteins. Connect Tissue Res. 2010; 51:230–9.

38. Matsui-Yuasa I, Otani S, Morisawa S, Takigawa M, Enomoto M, Suzuki F. Induction of spermidine/spermine N1-acetyltransferase by parathyroid hormone in rabbit costal chondrocytes in culture. J Biochem. 1985; 97:387–90.

39. Amraei M, Jia Z, Reboul P, Nabi IR. Acid-induced conformational changes in phosphoglucose isomerase result in its increased cell surface association and deposition on fibronectin fibrils. J Biol Chem. 2003; 278:38935–41.

40. Vaillancourt F, Fahmi H, Shi Q, Lavigne P, Ranger P, Fernandes JC, et al. 4-Hydroxynonenal induces apoptosis in human osteoarthritic chondrocytes: the protective role of glutathione-S-transferase. Arthritis Res Ther. 2008; 10:R107.

41. Grimmer C, Balbus N, Lang U, Aigner T, Cramer T, Müller L, et al. Regulation of type II collagen synthesis during osteoarthritis by prolyl-4-hydroxylases: possible influence of low oxygen levels. Am J Pathol. 2006; 169:491–502.
42. Iwamoto M, Jikko A, Murakami H, Shimazu A, Nakashima K, Iwamoto M, et al. Changes in parathyroid hormone receptors during chondrocyte cytodifferentiation. J Biol Chem. 1994; 269:17245–51.

43. Sanchez C, Deberg MA, Piccardi N, Msika P, Reginster JY, Henrotin YE. Subchondral bone osteoblasts induce phenotypic changes in human osteoarthritic chondrocytes. Osteoarthritis Cartilage. 2005; 13:988–97.

44. Mizuta H, Kudo S, Nakamura E, Takagi K, Hiraki Y. Expression of the PTH/PTHrP receptor in chondrogenic cells during the repair of full-thickness defects of articular cartilage. Osteoarthritis Cartilage. 2006; 14:944–52.

45. Guo J, Chung UI, Yang D, Karsenty G, Bringhurst FR, Kronenberg HM. PTH/PTHrP receptor delays chondrocyte hypertrophy via both Runx2-dependent and -independent pathways. Dev Biol. 2006; 292:116–28.

46. Meredith D, Gehl KA, Seymour J, Ellory JC, Wilkins RJ. Characterization of sulphate transporters in isolated bovine articular chondrocytes. J Orthop Res. 2007; 25:1145–53.

47. Rossi F, MacLean HE, Yuan W, Francis RO, Semenova E, Lin CS, et al. p107 and p130 Coordinately regulate proliferation, Cbfa1 expression, and hypertrophic differentiation during endochondral bone development. Dev Biol. 2002; 247:271–85.

48. Laplantine E, Rossi F, Sahni M, Basilico C, Cobrinik D. FGF signaling targets the pRb-related p107 and p130 proteins to induce chondrocyte growth arrest. J Cell Biol. 2002; 158:741–50.

49. Hummert TW, Schwartz Z, Sylvia VL, Dean DD, Boyan BD. Stathmin levels in growth plate chondrocytes are modulated by vitamin D3 metabolites and transforming growth factor-beta1 and are associated with proliferation. Endocrine. 2001; 15:93–101.
50. Selleri L, Depew MJ, Jacobs Y, Chanda SK, Tsang KY, Cheah KS, et al. Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development. 2001; 128:3543–57.

51. Vinall RL, Lo SH, Reddi AH. Regulation of articular chondrocyte phenotype by bone morphogenetic protein 7, interleukin 1, and cellular context is dependent on the cytoskeleton. Exp Cell Res. 2002; 272:32–44.

52. Singh MK, Petry M, Haenig B, Lescher B, Leitges M, Kispert A. The T-box transcription factor Tbx15 is required for skeletal development. Mech Dev. 2005; 122:131–44.

53. Polur I, Lee PL, Servais JM, Xu L, Li Y. Role of HTRA1, a serine protease, in the progression of articular cartilage degeneration. Histol Histopathol. 2010; 25:599–608.
54. Chamberland A, Wang E, Jones AR, Collins-Racie LA, LaVallie ER, Huang Y, et al. Identification of a novel HtrA1-susceptible cleavage site in human aggrecan: evidence for the involvement of HtrA1 in aggrecan proteol-ysis in vivo. J Biol Chem. 2009; 284:27352–9.
55. Krejci P, Prochazkova J, Smutny J, Chlebova K, Lin P, Aklian A, et al. FGFR3 signaling induces a reversible senescence phenotype in chondrocytes similar to oncogene-induced premature senescence. Bone. 2010; 47:102–10.

56. Kim JS, Ryoo ZY, Chun JS. Cytokine-like 1 (Cytl1) regulates the chondrogenesis of mesenchymal cells. J Biol Chem. 2007; 282:29359–67.

57. Massip L, Ectors F, Deprez P, Maleki M, Behets C, Lengelé B, et al. Expression of Hoxa2 in cells entering chondrogenesis impairs overall cartilage development. Differentiation. 2007; 75:256–67.

58. Mathy-Hartert M, Hogge L, Sanchez C, Deby-Dupont G, Crielaard JM, Henrotin Y. Interleukin-1beta and interleukin-6 disturb the antioxidant enzyme system in bovine chondrocytes: a possible explanation for oxidative stress generation. Osteoarthritis Cartilage. 2008; 16:756–63.
59. Kolupaeva V, Laplantine E, Basilico C. PP2A-mediated dephosphorylation of p107 plays a critical role in chondrocyte cell cycle arrest by FGF. PLoS One. 2008; 3:e3447.

60. Parker WL, Goldring MB, Philip A. Endoglin is expressed on human chondrocytes and forms a heteromeric complex with betaglycan in a ligand and type II TGFbeta receptor independent manner. J Bone Miner Res. 2003; 18:289–302.
61. Church V, Yamaguchi K, Tsang P, Akita K, Logan C, Francis-West P. Expression and function of Bapx1 during chick limb development. Anat Embryol (Berl). 2005; 209:461–9.

62. Konttinen YT, Mandelin J, Li TF, Salo J, Lassus J, Liljeström M, et al. Acidic cysteine endoproteinase cathepsin K in the degeneration of the superficial articular hyaline cartilage in osteoarthritis. Arthritis Rheum. 2002; 46:953–60.

63. Morko JP, Sö derström M, Sää mänen AM, Salminen HJ, Vuorio EI. Up regulation of cathepsin K expression in articular chondrocytes in a transgenic mouse model for osteoarthritis. Ann Rheum Dis. 2004; 63:649–55.

64. Connor JR, LePage C, Swift BA, Yamashita D, Bendele AM, Maul D, et al. Protective effects of a cathepsin K inhibitor, SB-553484, in the canine partial medial meniscectomy model of osteoarthritis. Osteoarthritis Cartilage. 2009; 17:1236–43.

65. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005; 120:15–20.

66. Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell- type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci U S A. 2006; 103:2746–51.
67. Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005; 310:1817–21.

68. Kobayashi T, Lu J, Cobb BS, Rodda SJ, McMahon AP, Schipani E, et al. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci U S A. 2008; 105:1949–54.

69. Stanczyk J, Pedrioli DM, Brentano F, Sanchez-Pernaute O, Kolling C, Gay RE, et al. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008; 58:1001–9.

70. Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006; 103:12481–6.
71. Luo X, Tsai LM, Shen N, Yu D. Evidence for micro-RNA-mediated regulation in rheumatic diseases. Ann Rheum Dis. 2010; 69(Suppl 1):i30–6.

72. Nakasa T, Miyaki S, Okubo A, Hashimoto M, Nishida K, Ochi M, et al. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum. 2008; 58:1284–92.

73. Yamasaki K, Nakasa T, Miyaki S, et al. Expression of MicroRNA-146a in osteoarthritis cartilage. Arthritis Rheum. 2009; 60:1035–41.

74. DelCarlo M, Loeser RF. Chondrocyte cell death mediated by reactive oxygen species-dependent activation of PKC-betaI. Am J Physiol Cell Physiol. 2006; 290:C802–11.
75. Saydam O, Shen Y, Würdinger T, Senol O, Boke E, James MF, et al. Downregulated microRNA-200a in meningiomas promotes tumor growth by reducing E-cadherin and activating the Wnt/beta-catenin signaling pathway. Mol Cell Biol. 2009; 29:5923–40.
76. Chun JS, Oh H, Yang S, Park M. Wnt signaling in cartilage development and degeneration. BMB Rep. 2008; 41:485–94.

77. Iliopoulos D, Malizos KN, Oikonomou P, Tsezou A. Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS One. 2008; 3:e3740.

78. Jones SW, Watkins G, Le Good N, Roberts S, Murphy CL, Brockbank SM, et al. The identification of differentially expressed microRNA in osteoarthritic tissue that modulate the production of TNF-alpha and MMP13. Osteoarthritis Cartilage. 2009; 17:464–72.
Figure 1.
Real-time RT-PCR results for miR-200a in chondrocytes in response to IL-1β. Total RNA was isolated from IL-1β treated monolayer chondrocytes obtained from donors different from those that were used in miRNA microarray. The RNA, U6 small nuclear 2 (RNU6-2) gene was used as a control to normalize differences in total RNA levels in each sample. The value of each control sample was set at 100 and was used to calculate the fold change in IL-1β treated chondrocytes. Data are from triplicate experiments using chondrocytes from 3 different donors. ∗denoted p<0.05 compared to control by using Mann-Whitney U test.
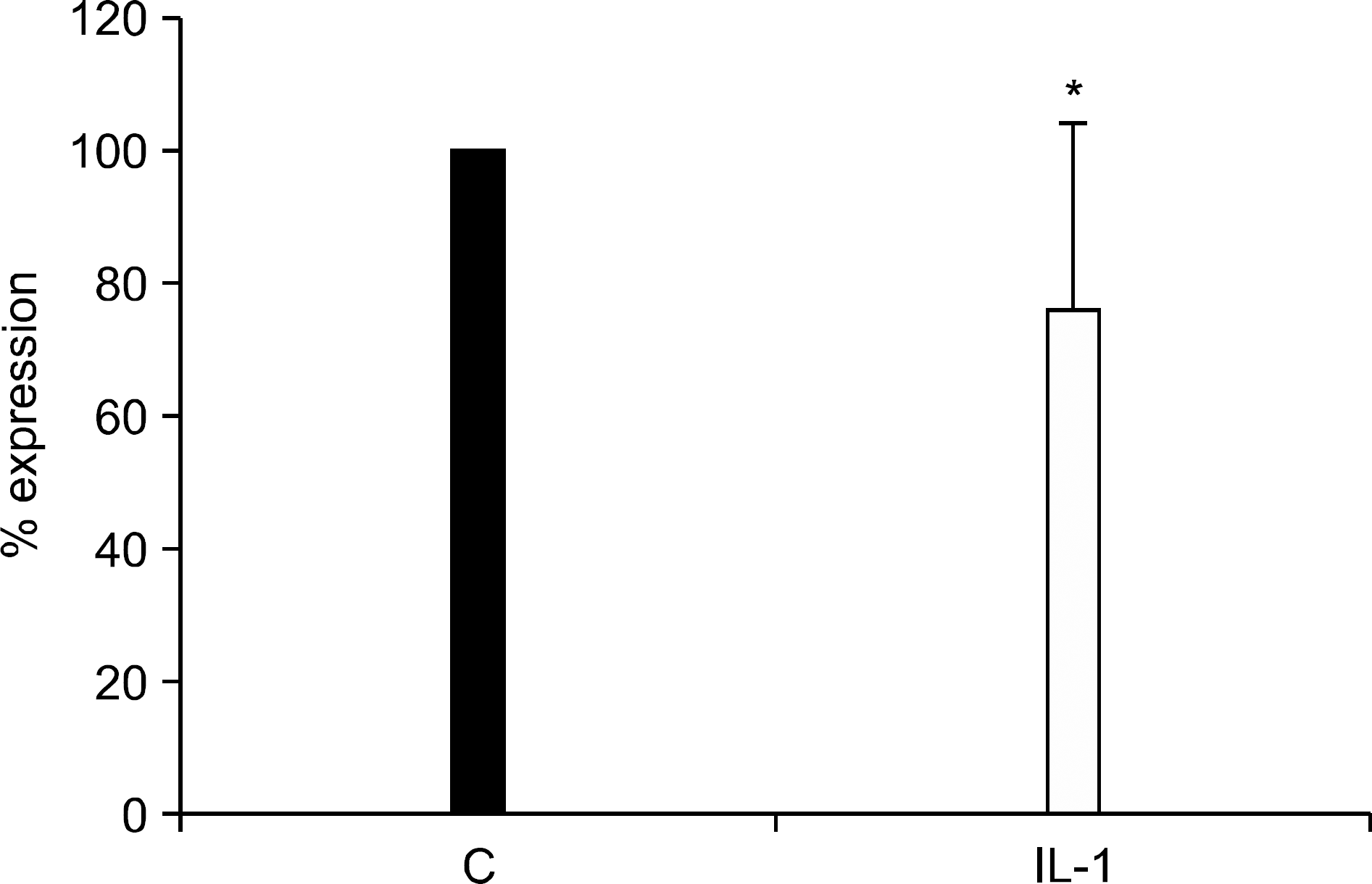
Figure 2.
RT-PCR results for tensin-1 and Tbx-15 in chondrocytes in response to IL-1β. Total RNA was isolated from IL-1β treated monolayer chondrocytes obtained from donors different from those that were used in mRNA microarray. GAPDH gene was used as a control to normalize differences in total RNA levels in each sample. Data are representative of duplicate experiments using chondrocytes from 3 different donors.
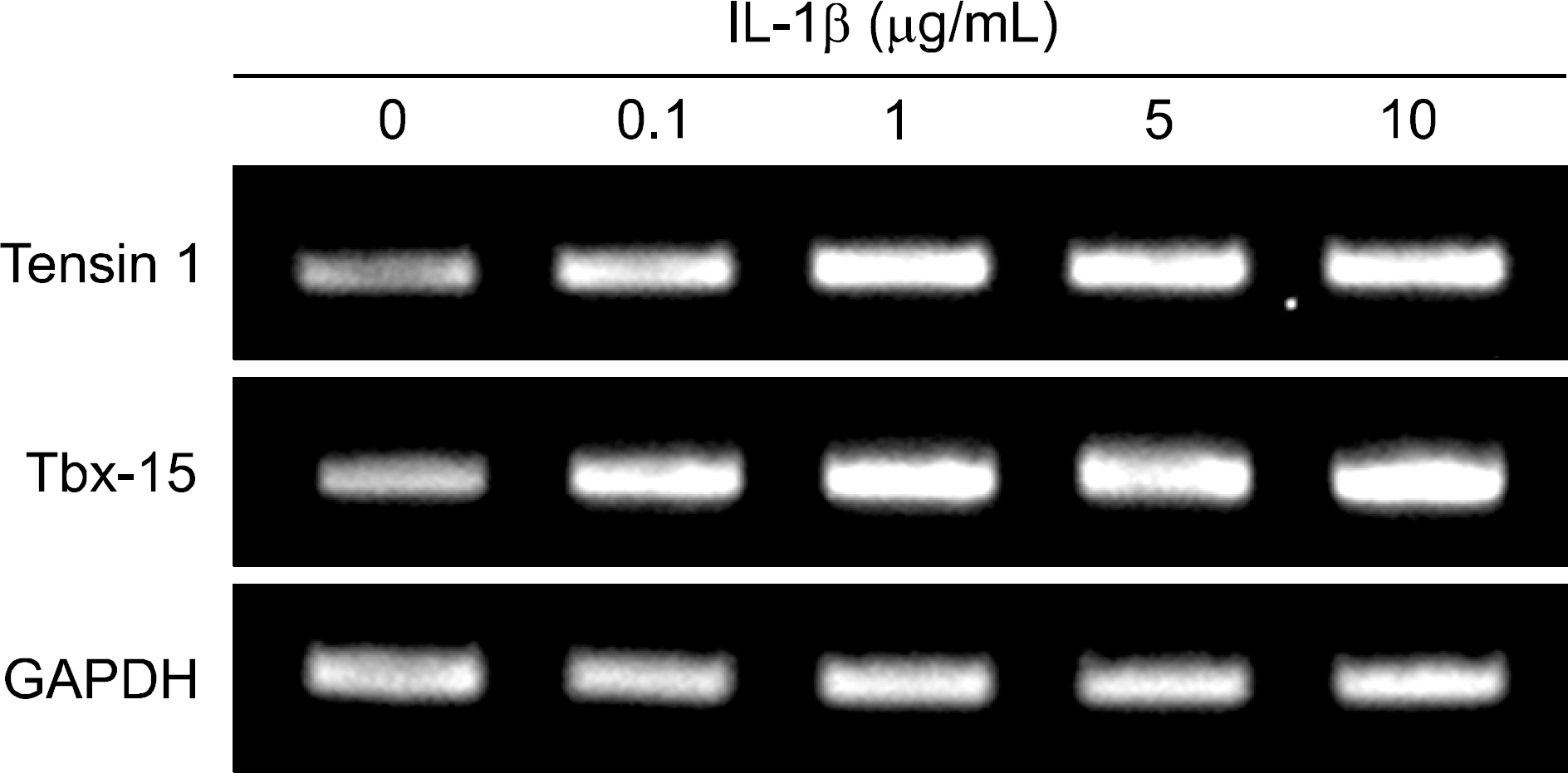
Figure 3.
In situ hybridization for miR-200a in normal and OA cartilages. Cartilage tissues obtained from OA patients at the time of joint replacement surgery were obtained from both lesion and non-lesion areas (defined as areas with preserved cartilage surface and thickness). For normal cartilage, macro and microscopically normal cartilage samples obtained from the femoral head of patients with femoral neck fracture were used. Data are representative of specimens from 4 different donors each for normal and OA (×100, original magnification).
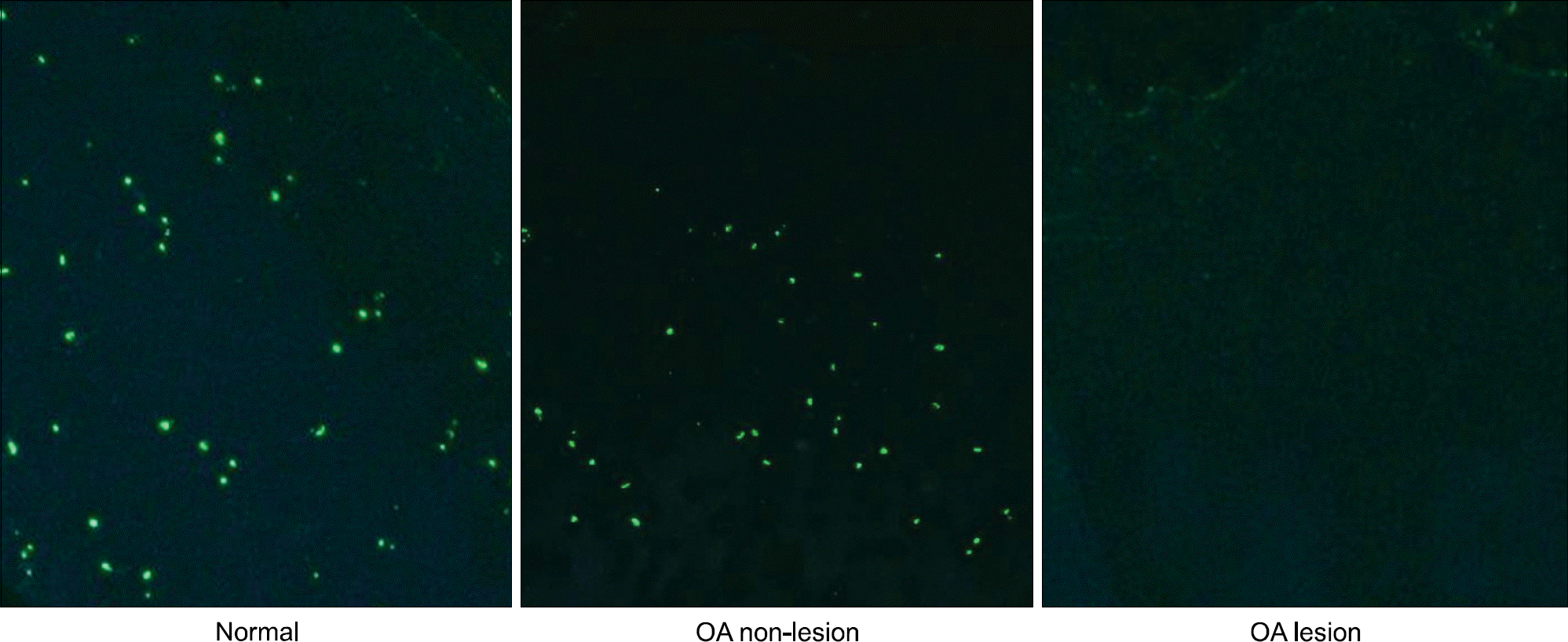
Table 1.
MiRNAs with differential expression between IL-1β treated and control chondrocytes from OA cartilages (n=6)
Table 2.
MiRNAs upregulated by IL-1β and their target mRNAs identified by integrative analysis
Table 3.
MiRNAs downregulated by IL-1β and their target mRNAs identified by integrative analysis




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download