Abstract
Background
Several high-throughput gene analysis techniques - differential display PCR, suppression subtraction hybridization (SSH), serial analysis of gene expression (SAGE), and DNA microarray - have permitted transcriptome profiling to understand the molecular pathogenesis of multifactorial diseases. But these techniques are of no great utility regarding feasibility, reproducibility, cost, and the amount of material required for analysis. To establish more practical method for transcription factor transcriptome profiling, we combined degenerate reverse transcriptase-polymerase chain reaction (RT-PCR) and single strand conformational polymorphism (SSCP) technique.
Methods
We categorized 417 human/mouse transcription factor mRNA into 92 small groups according to homology with ClustalW method and established 92 degenerate RT-PCR including common motives of the 92 small groups with the software program of CODEHOP, Primer Premier, Amplify 1.2. Further analysis on the amplified PCR products was performed by SSCP. This system was applied for the evaluation of changes on transcription factor transcriptome of differentiated 3T3-L1 adipocyte treated with TNF-α.
Results
82 groups and 52 groups showed amplification of PCR before and after TNF-α treatment respectively and 24 groups showed significant amplification difference after TNF-α treatment. After TNF-α treatment for 48 hours, mRNA expressions of group 7, 30, and 33 which include adipocyte related transcription factors such as CEBP-α, RXR-α, PPAR-γ were downregulated and mRNA expression of group 8 including preadipocyte abundant CEBP-β was upregulated. These results are largely concordant with the results analyzed by oligonucleotide microarray. Randomly selected single PCR bands of group 28 and 75 on agarose electrophoresis displayed additional multiple bands by SSCP and necessitated addition of this technique to degenerate RT-PCR for further analysis.
Figures and Tables
Fig. 1
Effect of TNF-α on protein level of insulin receptor substrate 1 (IRS-1) in 3T3-L1 adipocytes by gel electrophoresis and immunoblotting (Ctrl: 3T3-L1 preadipocyte, lane 1: fully differentiated 3T3-L1 adipocyte, lane 2: adipocyte treated with 100 nM insulin for 15 min, lane 3: adipocyte treated with 3 ng/mL TNF-α for 48 h, lane 4: adipocyte treated with TNF-α and insulin).
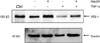
Fig. 2
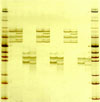
Agarose gel electrophoresis of transcription factor degenerate RT-PCRs on total RNAs of fully differentiated 3T3-L1 adipocytes before and after TNF-α treatment. Numbers above the columns mean categorized groups of transcription factors. Each left and right column below each number shows the result of RT-PCR from 3T3-L1 adipocytes before and after TNF-α treatment, respectively. Columns without number on the top are markers for the sizes (base pair) of PCR product.

Fig. 3
SSCP (single strand conformational polymorphism) analysis of transcription factor RT-PCRs. The products, taken from 28 and 75 RT-PCRs in Fig. 2 were denatured at 96℃ for 5min, cooled in ice for 5 min, and then 3 times loaded on 12% nondenaturing polyacrylamide gel.
References
1. Kiechle FL, Holland-Staley CA. Genomics, Transcriptomics, Proteomics, and Numbers. Arch Pathol Lab Med. 2003. 127:1089–1097.
2. Müller CW. Transcription Factors: global and detailed views. Curr Opin Struc Biol. 2001. 11:26–32.
3. Warren AJ. Eukaryotic transcription factors. Curr Opin Struc Biol. 2002. 12:107–114.
4. Connolly SB, Sadlier D, Kieran NE, Doran P, Brady HR. Transcriptome profiling and the pathogenesis of diabetic complications. J Am Soc Nephrol. 2003. 14:S279–S283.
5. Permana PA, Del Parigi A, Tataranni PA. Microarray gene expression profiling in obesity and insulin resistance. Nutrition. 2004. 20:134–138.
6. Viguerie N, Poitou C, Cancello R, Stich V, Clément K, Langin D. Transcriptomics applied to obesity and caloric restriction. Biochimie. 2005. 87:117–123.
7. Heinemeyer T, Chen X, Karas H, Kel AE, Kel OV, Liebich I, Meinhardt T, Reuter I, Schacherer F, Wingender E. Expanding the TRANSFAC database towards an expert system of regulatory molecular mechanisms. Nucleic Acids Res. 1999. 27:318–322.
8. Wingender E, Chen X, Hehl R, Karas H, Liebuch I, Matys V, Meinhardt T, Prϋβ M, Reuter I, Schacherer F. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 2000. 28:316–319.
9. Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 1998. 26:1628–1635.
10. Rose TM, Henikoff JG, Henikoff S. CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 2003. 31:3763–3766.
11. Cuchacovich R. Clinical Applications of the polymerase chain reaction : An update. Infect Dis Clin North Am. 2006. 20:735–758.
12. Krajewski KM, Shy ME. Genetic testing in neuromuscular disease. Neurol Clin. 2004. 22:481–508.
13. Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 1993. 259:87–91.
14. Hofmann C, Lorenz K, Braithwaite SS, Colca JR, Palazuk BJ, Hotamisligil GS, Spiegelman BM. Altered gene expression for tumor necrosis factor alpha and its receptors during drug and dietary modulation of insulin resistance. Endocrinology. 1994. 134:264–270.
15. Hamann A, Benecke H, Le Marchand-Brustel Y, Susulic VS, Lowell BB, Flier JS. Characterization of insulin resistance and NIDDM in transgenic mice with reduced brown fat. Diabetes. 1995. 44:1266–1273.
16. Hotamisligil GS, Arner P, Caro JF, Atkinson RL, Spiegelman BM. Increased adipose tissue expression of tumor necrosis factor-alpha in human obesity and insulin resistance. J Clin Invest. 1995. 95:2409–2415.
17. Kern PA, Saghizadeh M, Ong JM, Bosch RJ, Deem R, Simsolo RB. The expression of tumor necrosis factor in human adipose tissue: regulation by obesity, weight loss, and relationship to lipoprotein lipase. J Clin Invest. 1995. 95:2111–2119.
18. Green A, Dobias SB, Walters DJ, Brasier AR. Tumor necrosis factor increases the rate of lipolysis in primary cultures of adipocytes without altering levels of hormone-sensitive lipase. Endocrinology. 1994. 134:2581–2588.
19. Aguirre V, Werner ED, Giraud J, Lee YH, Shoelson SE, White MF. Phosphorylation of Ser307 in insulin receptor substrate-1 blocks interactions with the insulin receptor and inhibit insulin action. J Biol Chem. 2002. 277:1531–1537.
20. Guo D, Donner DB. Tumor necrosis factor promotes phosphorylation and binding of insulin receptor substrate 1 to phosphatidylinositol 3-kinase in 3T3-L1 adipocytes. J Biol Chem. 1996. 271:615–618.
21. Heart E, Choi WS, Sung CK. Glucosamine-induced insulin resistance in 3T3-L1 adipocytes. Am J Physiol Endocrinol Metab. 2000. 278:E103–E112.
22. Fasshauer M, Klein J, Neumann S, Eszlinger M, Paschke R. Tumor necrosis factor alphais a negative regulator of resistin gene expression and secretion in 3T3-L1 adipocytes. Biochem Biophys Res Commun. 2001. 288:1027–1031.
23. Garcia de Herreros A, Birnbaum M. The acquisition of increased insulin-responsive hexose transport in 3T3-L1 adipocytes correlates with expression of a novel transporter gene. J Biol Chem. 1989. 264:19994–19999.
24. Grimwade D, Du MC, Langabeer S, Rogers J, Solomon E. Screening for mutations of Bcl10 in leukaemia. Br J Haematol. 2000. 109:611–615.
25. Zhao W, Zhang L, Kobari Y, Misaki Y, Yamamoto K. Analysis of accumulating clonotypes of T cell in joints of a spontaneous murine model of rheumatoid arthritis. Cell Mol Immunol. 2004. 1:300–303.
26. Peraldi P, Xu M, Spiegelman BM. Thiazolidinediones block tumor necrosis factor-alpha-induced inhibition of insulin signaling. J Clin Invest. 1997. 100:1863–1869.
27. Hotamisligil GS, Peraldi P, Budavari A, Ellis R, White MF, Spiegelman BM. IRS-1-mediated inhibition of insulin receptor tyrosine kinase activity in TNF-alpha- and obesity-induced insulin resistance. Science. 1996. 271:665–668.
28. Stephens JM, Lee J, Pilch PF. Tumor necrosis factor-alpha-induced insulin resistance in 3T3-L1 adipocytes is accompanied by a loss of insulin receptor substrate-1 and GLUT4 expression without a loss of insulin receptor mediated signal transduction. J Biol Chem. 1997. 272:971–976.
29. Ruan H, Hacohen N, Golub TR, Van Parijs L, Lodish HF. Tumor necrosis factor-α suppresses adipocyte-specific genes and activates expression of preadipocyte genes in 3T3-L1 adipocytes: nuclear factor-kappaB activation by TNF-α is obligatory. Diabetes. 2002. 51:1319–1336.
30. Ruan H, Miles PDG, Ladd CM, Ross K, Golub TR, Olefsky JM, Lodish HF. Profiling gene transcription in vivo reveals adipose tissue as an immediate target of tumor necrosis factor-alpha: implications for insulin resistance. Diabetes. 2002. 51:3176–3188.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download