Abstract
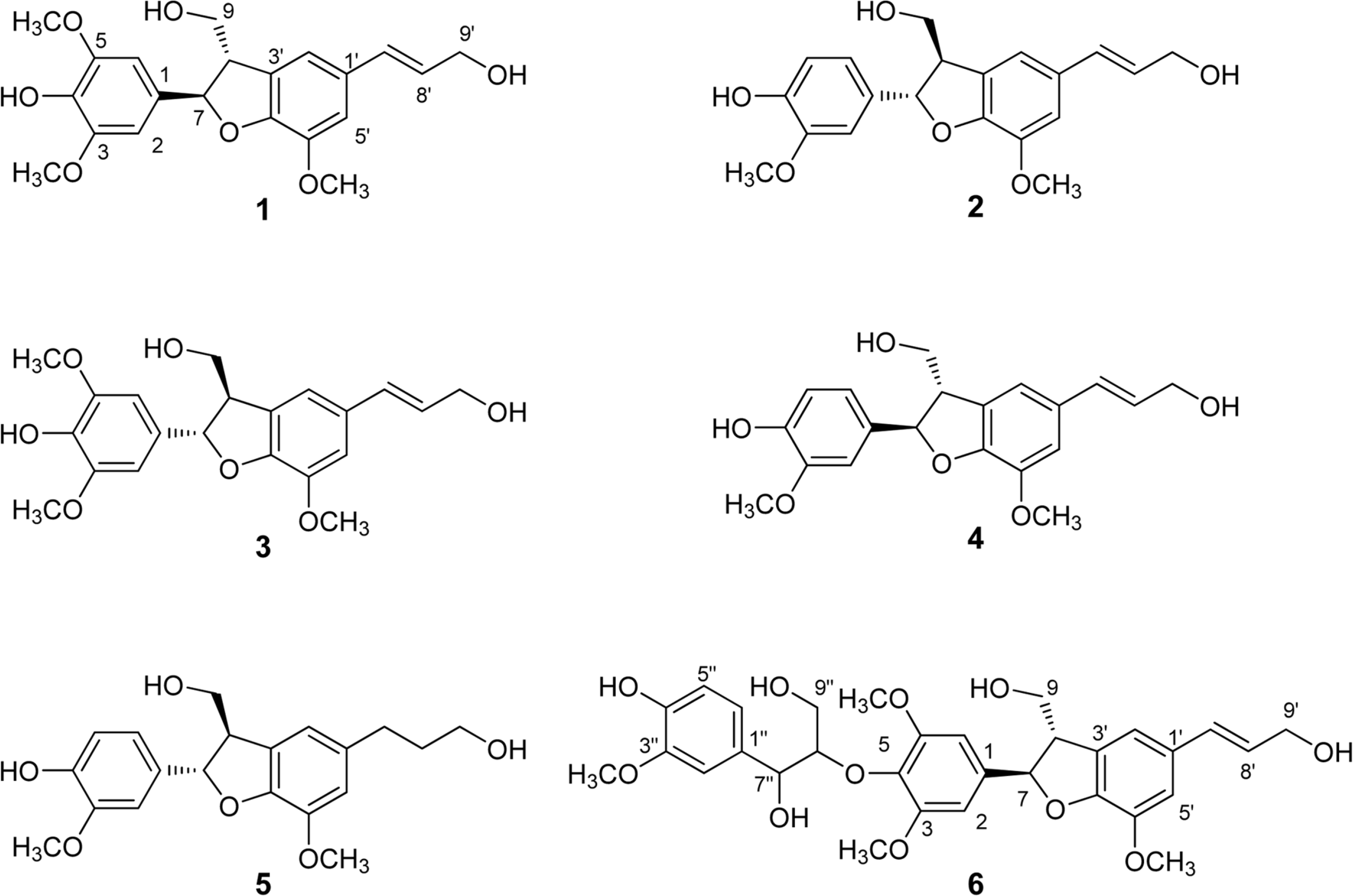
Anti-inflammatory effects of dihydrobenzofuran neolignans isolated from Euonymus alatus leaves and twigs were evaluated in lipopolysaccharide (LPS)-stimulated RAW264.7 macrophage cells. Six neolignans, (+)-simulanol (1), (+)-dehydrodiconiferyl alcohol (2), (−)-simulanol (3), (−)-dehydrodiconiferyl alcohol (4), (+)-dihydrodehyrodiconiferyl alcohol (5), threo-buddlenol B (6) effectively inhibited the production of nitric oxide (NO) induced by LPS, and the activity of iNOS. (−)-dehydrodiconiferyl alcohol (4), which showed the most potent inhibitory activity, attenuated the activity of iNOS enzyme and also the expression of iNOS and COX-2 proteins. The subsequent production of proinflammatory cytokines, interleukin-1β, interleukin-6, tumor necrosis factor-α and prostaglandin E2 were also inhibited by the pretreatment of RAW264.7 cells with (−)-dehydrodiconiferyl alcohol (4). These neolignans are thought to contribute to anti-inflammatory effects of E. alatus, and expected to be potential candidates to prevent/treat inflammation-related diseases.
Go to : 
REFERENCES
(1). Kaplanski G., Marin V., Montero-Julian F., Mantovani A., Farnarier C.Trends Immunol. 2003; 24:25–29.
(3). Jeong E. J., Yang H., Kim S. H., Kang S. Y., Sung S. H., Kim Y. C.Food Chem. Toxicol. 2011; 49:1394–1398.
(4). Jeong E. J., Cho J. H., Sung S. H., Kim S. Y., Kim Y. C.Bioorg. Med. Chem. Lett. 2011; 15:2283–2286.
(5). Akihisa T., Yamamoto K., Tamura T., Iida T., Nambara T., Chang F. C.Chem. Pharm. Bull. 1992; 40:789–791.
(6). De Fátima Silva G. D., Duarte L. P., Da Silva Paes H. C., De Sousa J. R., Nonato M. C., Portezani P. J., Mascarenhas Y. P. J.Braz. Chem. Soc. 1998; 9:461–464.
(7). Liu C. M., Wang H. X., Wei S. L., Gao K. J.Nat. Prod. 2008; 71:789–792.
(8). Fang J. M., Lee C. K., Cheng Y. S.Phytochemistry. 1992; 31:3659–3661.
(9). Yang Y. P., Cheng M. J., Teng C. M., Chang Y. L., Tsai I. L., Chen I. S.Phytochemistry. 2002; 61:567–572.
(10). Meng J., Jiang T., Bhatti H. A., Siddiqui B. S., Dixon S., Kilburn J. D.Org. Biomol. Chem. 2010; 8:107–113.
(11). Lourith N., Katayama T., Suzuki T. J.Wood Sci. 2005; 51:370–378.
(12). Matsuda S., Kadota S., Tai T., Kikuchi T.Chem. Pharm. Bull. 1984; 32:5066–5069.
(13). Dawson V. L., Brahmbhatt H. P., Mong J. A., Dawson T. M.Neuropharmacology. 1994; 33:1425–1430.
(14). Korhonen R., Lahti A., Kankaanranta H., Moilanen E.Curr. Drug Targets Inflamm. Allergy. 2005; 4:471–479.
(15). Yamashita T., Kawashima S., Ohashi Y., Ozaki M., Ueyama T., Ishida T., Inoue N., Hirata K., Akita H., Yokoyama M.Circulation. 2000; 101:931–937.
(16). Penglis P. S., Cleland L. G., Demasi M., Caughey G. E., James M. J. J.Immunol. 2000; 165:1605–1611.
(18). Marletta M. A. J.Biol. Chem. 1993; 268:12231–12234.
(19). Duval D. L., Miller D. R., Collier J., Billings R. E.Mol. Pharmacol. 1996; 50:277–284.
(20). Son H. J., Lee H. J., Yun-Choi H. S., Ryu J. H.Planta Med. 2000; 66:469–471.
(21). Chen T. H., Kao Y. C., Chen B. C., Chen C. H., Chan P., Lee H. M.Eur. J. Pharmacol. 2006; 541:138–146.
(22). Hamasaki Y., Kobayashi I., Zaitu M., Tsuji K., Kita M., Hayasaki R., Muro E., Yamamoto S., Matsuo M., Ichimaru T., Miyazaki S.Planta Med. 1999; 65:222–226.
(23). Wang J. P., Raung S. L., Chen C. C., Kuo J. S., Teng C. M.Naunyn Schmiedebergs Arch. Pharmacol. 1993; 348:663–669.
(24). Oh J. H., Kang L. L., Ban J. O., Kim Y. H., Kim K. H., Han S. B., Hong J. T.Chem. Biol. Interact. 2009; 180:506–514.
(25). Choi M. S., Lee S. H., Cho H. S., Kim Y., Yun Y. P., Jung H. Y., Jung J. K., Lee B. C., Pyo H. B., Hong J. T.Eur. J. Pharmacol. 2007; 556:181–189.
(26). Connell L., Mclnnes I. B.Best Pract. Res. Clin. Rheumatol. 2006; 20:865–878.
(27). Aggarwal B. B., Natarajan K.Eur. Cytokine Netw. 1996; 7:93–124.
Go to : 
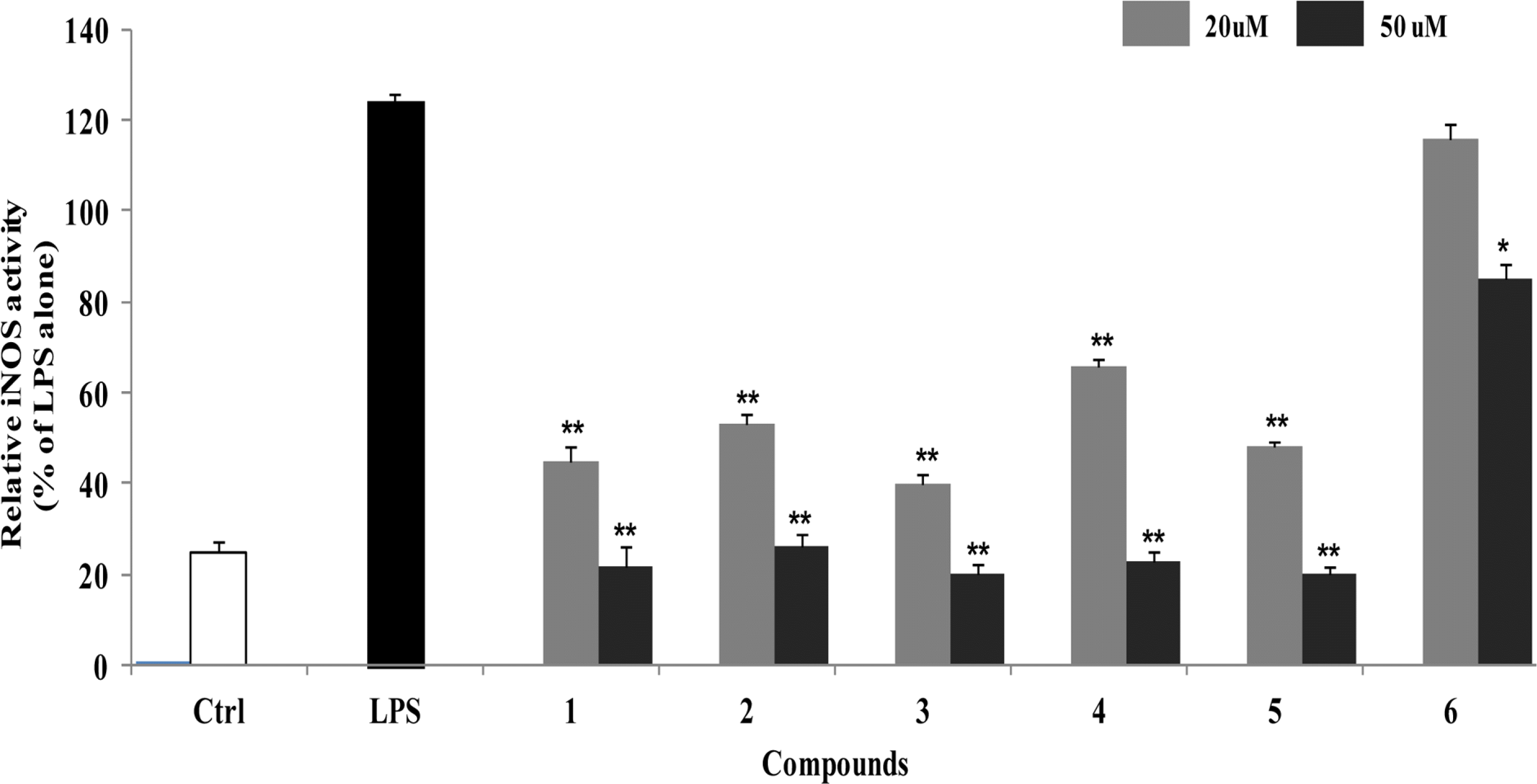 | Fig. 2.Inhibitory effects of compounds 1–6 on iNOS enzyme activity in LPS-stimulated RAW264.7 cells. RAW264.7 cells were treated with compounds 1–6 for 1 h before exposure to LPS for 24 h. The fluorescent product was assessed as described in the Materials and Methods. The values shown are the mean ± s.d. of data from three independent experiments. Results differ significantly from LPS alone,∗P < 0.01,∗∗P < 0.001. |
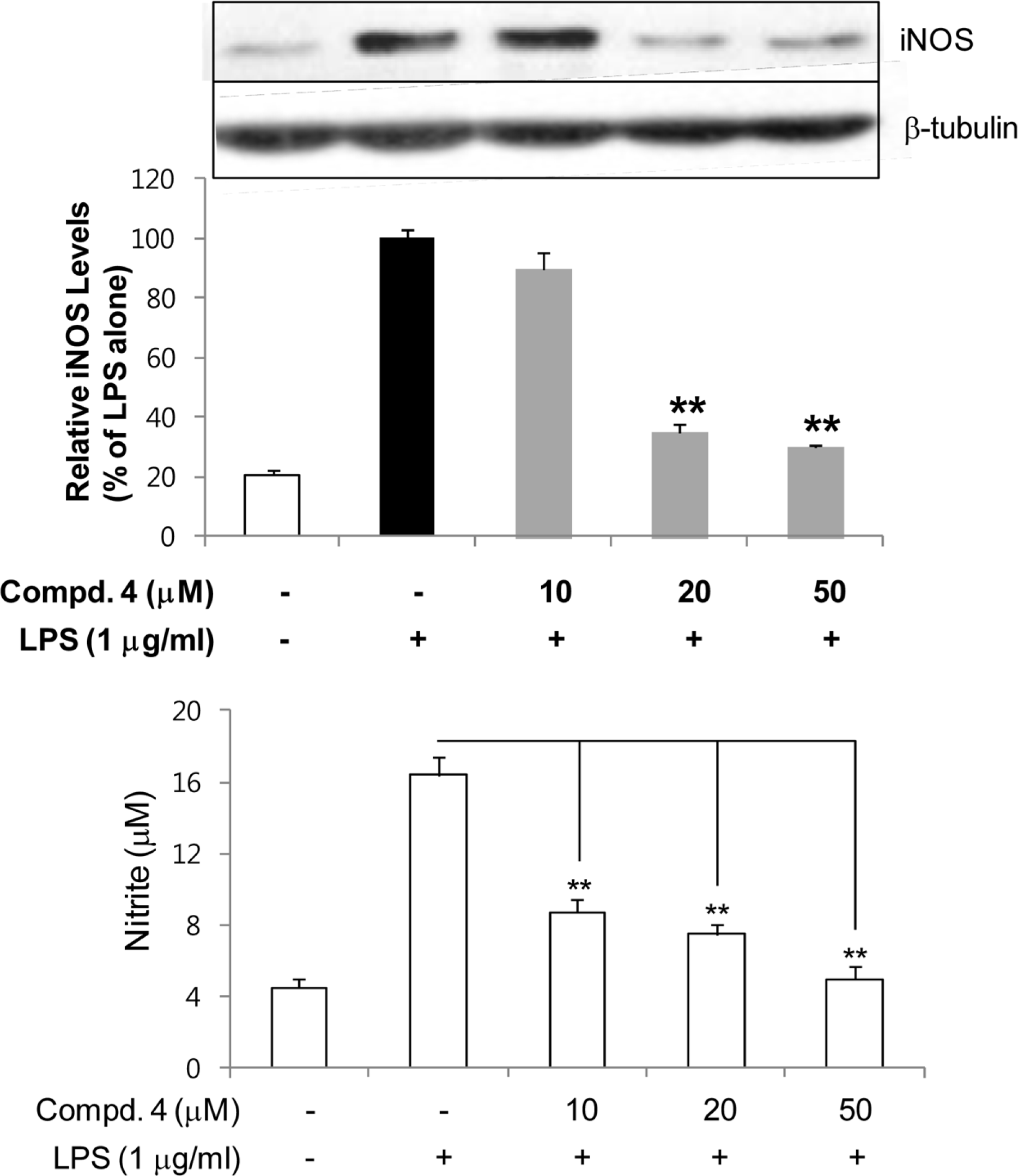 | Fig. 3.Inhibitory effects of compounds 4 on the expression of iNOS protein in LPS-stimulated RAW264.7 cells. RAW264.7 cells were treated with compound 4 for 1 h, and then exposed to LPS for 24 h. Cell lysates (40 ug protein) were prepared and subjected to Western blot analysis using iNOS-specific antibodies. The relative protein levels were quantified by scanning densi-tometry and normalized to b-tubulin protein. NO production was measured by the Griess reaction and sodium nitrite was used as a standard. The values shown are the mean ± s.d. of data from three independent experiments. Results differ significantly from LPS alone,∗P < 0.01,∗∗P < 0.001. |
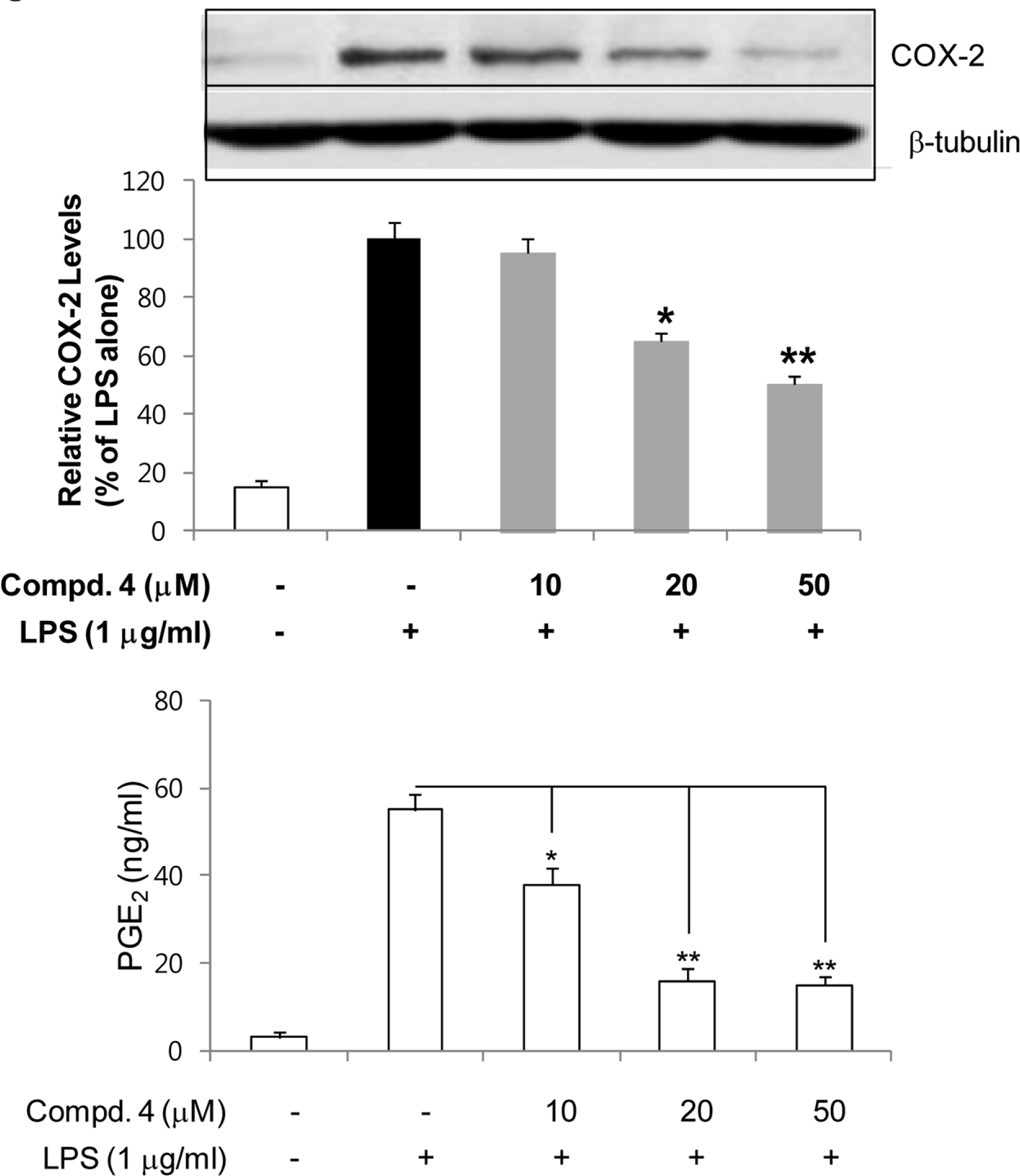 | Fig. 4.Inhibitory effects of compounds 4 on the expression of COX-2 protein in LPS-stimulated RAW264.7 cells. RAW264.7 cells were treated with compound 4 for 1 h, and then exposed to LPS for 24 h. Cell lysates (40 ug protein) were prepared and subjected to Western blot analysis using COX-2-specific antibodies. The relative protein levels were quantified by scanning densito-metry and normalized to b-tubulin protein. The concentrations of PGE2 in the culture medium were determined using ELISA system as described in Materials and Methods. The values shown are the mean ± s.d. of data from three independent experiments. Results differ significantly from LPS alone,∗P <0.01,∗∗P <0.001. |
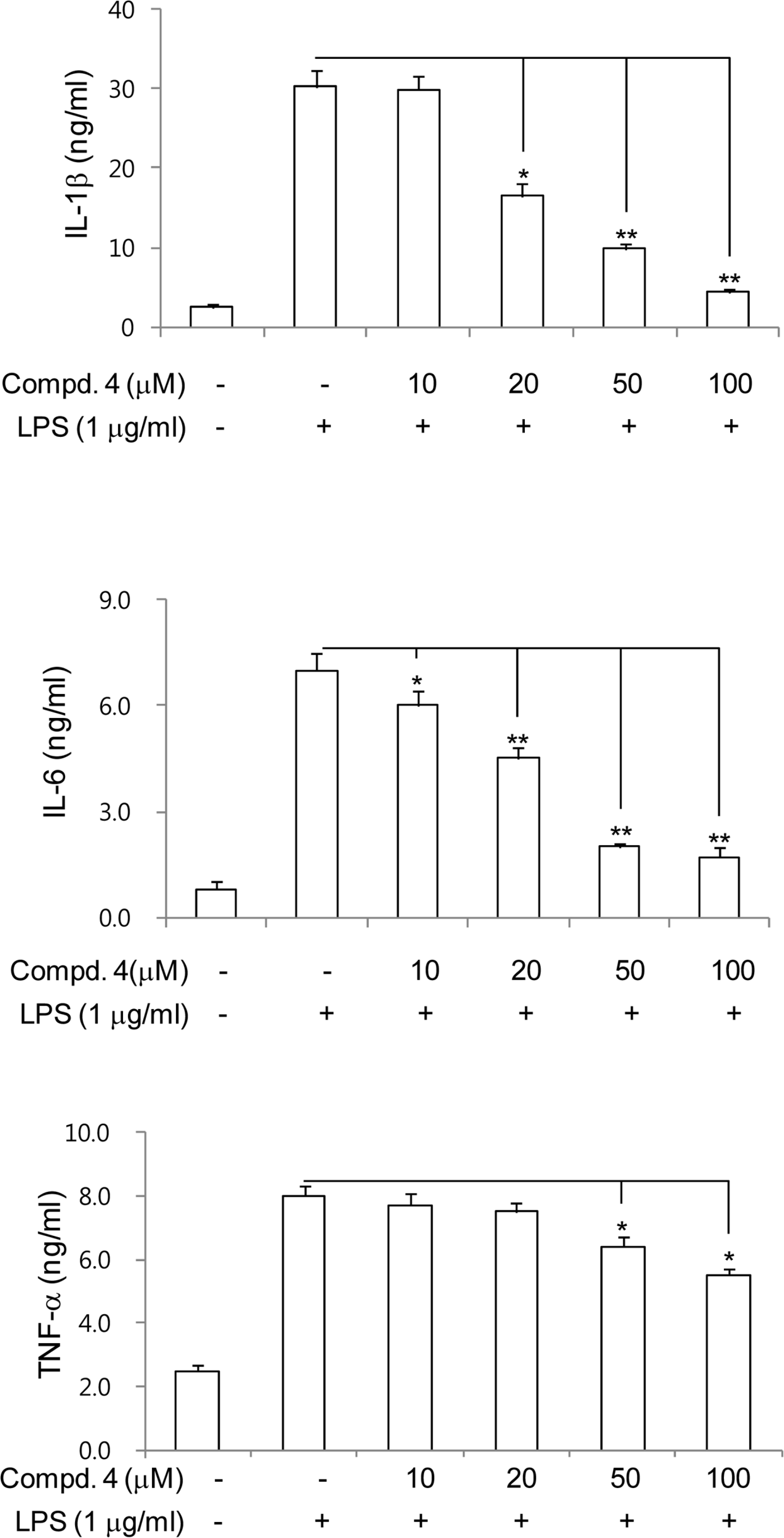 | Fig. 5.Inhibitory effects of compounds 4 on the production of proinflammatory cytokines in LPS-stimulated RAW264.7 cells. RAW264.7 cells were treated with compound 4 for 1 h, and then exposed to LPS for 24 h. The concentrations of IL-1β, IL-6 and TNF-α in the culture medium were determined using ELISA system as described in Materials and Methods. The values shown are the mean ± s.d. of data from three independent experiments. Results differ significantly from LPS alone,∗P <0.01,∗∗P <0.001. |
Table 1.
Inhibitory effects of compounds 1–6 isolated from E. alatus leaves and twigs on NO production induced by LPS in RAW264.7 cells
| Concentration | 10 μ M | 20 μ M | 50 μ M | 100 μ M | IC50 μ M |
|---|---|---|---|---|---|
| Nitrite (μ M) | |||||
| Control | 5.1 ± ± 0.2 | ||||
| LPS | 56.5 ±0.3 | ||||
| 1 | 33.2 ± 1.9∗ | 20.2 ± 1.4∗ | 18.8 ± 2.7∗ | 15.4 ± 0.8∗∗ | 11.7 ± 1.2 |
| 2 | 29.3 ± 2.3∗ | 23.2 ± 1.6∗ | 11.2 ± 1.2∗∗ | 66.1 ± 0.1∗∗ | 9.3 ± 1.4 |
| 3 | 32.9 ± 1.7∗ | 26.4 ± 0.3∗ | 17.3 ± 1.5∗∗ | 14.9 ± 0.5∗∗ | 12.8 ± 2.0 |
| 4 | 26.6 ± 2.0∗ | 21.4 ± 1.5∗ | 68.7 ± 0.2∗∗ | 62.9 ± 0.1∗∗ | 8.5 ± 0.8 |
| 5 | 31.7 ± 1.5∗ | 24.2 ± 1.2∗ | 12.7 ± 0.8∗∗ | 66.9 ± 0.2∗∗ | 9.8 ± 2.0 |
| 6 | 48.8 ± 2.5 | 31.9 ± 0.9∗ | 29.8 ± 1.3∗ | 28.9 ± 1.0∗ | 21.3 ± 3.3 |
| L-NAME | 48.5 ± 2.3 | ||||
| L-NNA | 60.3 ± 1.7 | ||||
| L-NMMA | 32.9 ± 2.2 | ||||




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download