Abstract
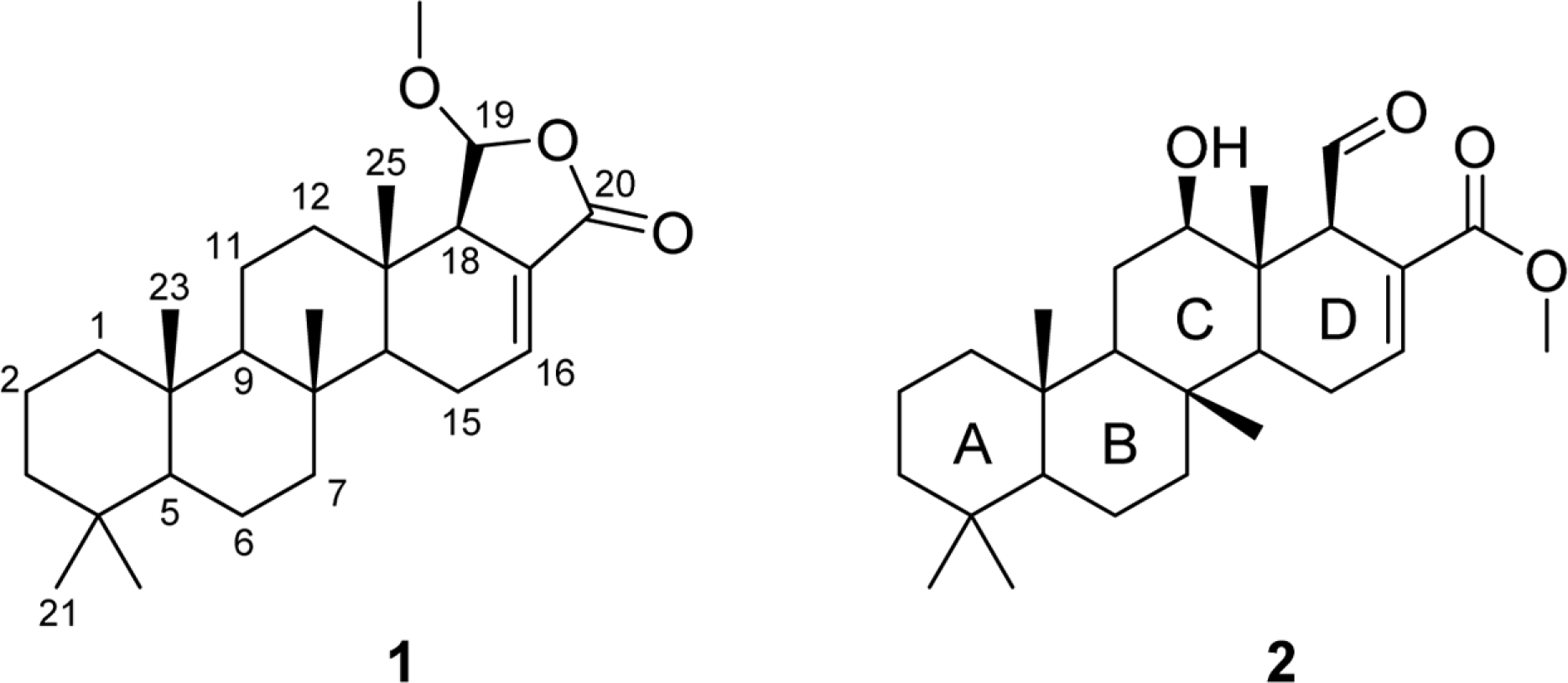
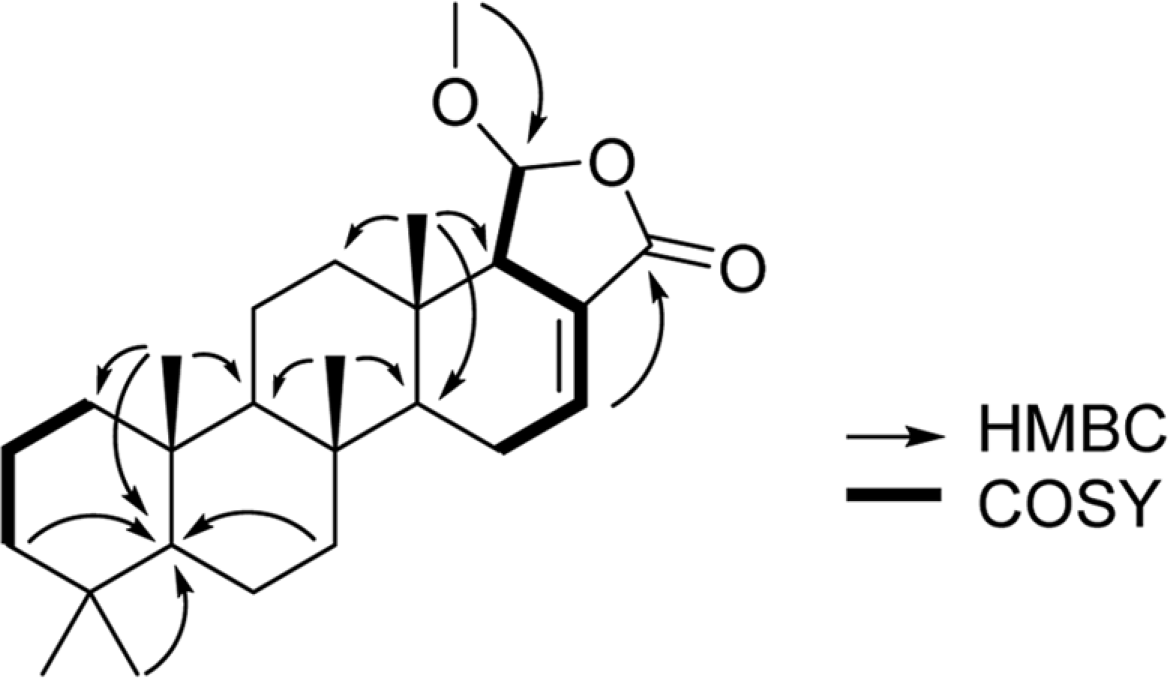
Intensive chemical investigation of Korean marine sponge Spongia sp. has led to the isolation of two new scalaranes. The planar structures of the new compounds 1 and 2 were determined through 1D and 2D NMR spectral data analysis, while the relative stereochemistry of the compounds was determined based on the analysis of 1H-1H coupling constants and NOESY spectroscopic data. Compounds 1 and 2 did not display any significant biological activities on farnesoid X-activated receptor (FXR) in co-transfection assay.
REFERENCES
(1). Fattorusso E., Gerwick W. H., Taglialatela-Scafati O.Handbook of Marine Natural Products. Springer;New York: 2012. p. 4.
(2). Wang L., Yang B., Lin X. P., Zhou X. F., Liu Y.Nat. Prod. Rep. 2013; 30:455–473.
(3). González M. A.Curr. Bioact. Compd. 2010; 6:178–206.
(4). Fattorusso E., Magno S., Santacroce C., Sica D.Tetrahedron. 1972; 28:5993–5997.
(5a). Harinantenaina L., Brodie P. J., Maharavo J., Bakary G., TenDyke K., Shen Y., Kingston D. G.Bioorg. Med. Chem. 2013; 21:2912–2917.
(5b). Hassan M. H. A., Rateb M. E., Hetta M., Abdelaziz T. A., Sleim M. A., Jaspars M., Mohammed R.Tetrahedron. 2015; 71:577–583.
(5c). Yong K. W., Mudianta I. W., Cheney K. L., Mollo E., Blanchfield J. T., Garson M. J. J.Nat. Prod. 2015; 78:421–430.
(6a). Cimino G., De Rosa S., De Stefano S., Sodano G.Comp. Biochem. Phys. B. 1982; 73:471–474.
(6b). Rogers S. D., Paul V. J.Mar. Ecol. Prog. Ser. 1991; 77:221–232.
(6c). Avila C., Paul V. J.Mar. Ecol. Prog. Ser. 1997; 150:171–180.
(6d). Walker R. P., Thompson J. E., Faulkner D. J.J. Org. Chem. 1980; 45:4976–4979.
(6e). Thompson J. E., Walker R. P., Faulkner D. J.Mar. Biol. 1985; 88:11–21.
(7). Sera Y., Adachi K., Shizuri Y. J.Nat. Prod. 1999; 62:152–154.
(8a). Diyabalanage T., Ratnayake R., Bokesch H. R., Ransom T. T., Henrich C. J., Beutler J. A., McMahon J. B., Gustafson K. R.J. Nat. Prod. 2012; 75:1490–1494.
(8b). Cao F., Wu Z. H., Shao C. L., Pang S., Liang X. Y., de Voogd N. J., Wang C. Y.Org. Biomol. Chem. 2015; 13:4016–4024.
(9). Chang Y. C., Tseng S. W., Liu L. L., Chou Y., Ho Y. S., Lu M. C., Su J. H.Mar. Drugs. 2012; 10:987–997.
(10). Mo S., Krunic A., Pegan S. D., Franzblau S. G., Orjala J. J.Nat. Prod. 2009; 72:2043–2045.
(11). Marshall L. A., Winkler J. D., Griswold D. E., Bolognese B., Roshak A., Sung C. M., Webb E. F., Jacobs R. J.Pharmacol. Exp. Ther. 1994; 268:709–717.
(12). Festa C., Cassiano C., D'Auria M. V., Debitus C., Monti M. C., De Marino S.Org. Biomol. Chem. 2014; 12:8646–8655.
(13). Abdjul D. B., Yamazaki H., Takahashi O., Kirikoshi R., Mangindaan R. E., Namikoshi M.Bioorg. Med. Chem. Lett. 2015; 25:904–907.
(14). Putra M. Y., Bavestrello G., Cerrano C., Renga B., D'Amore C., Fiorucci S., Fattorusso E., Taglialatela-Scafati O.Steroids. 2012; 77:433–440.
(15a). Jeon J. E., Bae J., Lee K. J., Oh K. B., Shin J.J. Nat. Prod. 2011; 74:847–851.
(15b). Lee Y. J., Lee J. W., Lee D. G., Lee H. S., Kang J. S., Yun J.Int. J. Mol. Sci. 2014; 15:20045–20053.
(16a). Nam S. J., Ko H., Shin M., Ham J., Chin J., Kim Y., Kim H., Shin K., Choi H., Kang H.Bioorg. Med. Chem. Lett. 2006; 16:5398–5402.
(16b). Nam S. J., Ko H., Ju M. K., Hwang H., Chin J., Ham J., Lee B., Lee J., Won D. H., Choi H., Ko J., Shin K., Oh T., Kim S., Rho J. R., Kang H. J.Nat. Prod. 2007; 70:1691–1695.
(16c). Hahn D., Won D. H., Mun B., Kim H., Han C., Wang W., Chun T., Park S., Yoon D., Choi H., Nam S. J., Ekins M., Chin J., Kang H.Bioorg. Med. Chem. Lett. 2013; 23:2336–2339.
Table 1.
1H and 13C NMR data of 1 and 2 in CDCl3.
| 1a | 2b | |||||
|---|---|---|---|---|---|---|
| No. | δC, mc | δH, m, J (Hz) | COSY | HMBC | δC, mc | δH, m, J (Hz) |
| 1 | 40.1, CH2 | 0.79, m | 2 | 3, 23 | 40.4, CH2 | 0.76, m |
| 1.67, m | 1.67, m | |||||
| 2 | 18.8, CH2 | 1.51, m | 1, 3 | 18.7, CH2 | 1.38, m | |
| 1.55, m | 1.48, m | |||||
| 3 | 42.2, CH2 | 1.13, dt (9.4, 3.7) | 2 | 5 | 42.2, CH2 | 1.15, m |
| 1.39, m | 1.38, m | |||||
| 4 | 33.5, C | 33.4, C | ||||
| 5 | 56.6, CH | 0.78, m | 56.5, CH | 0.76, m | ||
| 6 | 18.2, CH2 | 1.40, m | 18.2, CH2 | 1.33, m | ||
| 1.51, m | 1.48, m | |||||
| 7 | 41.8, CH2 | 0.96, dt (13.2, 3.5) | 5, 9, 24 | 41.8, CH2 | 0.89, m | |
| 1.69, d (10.2) | 1.69, m | |||||
| 8 | 37.8, C | 37.3, C | ||||
| 9 | 61.4, CH | 0.85, m | 58.5, CH | 0.83, m | ||
| 10 | 37.8, C | 37.3, C | ||||
| 11 | 17.3, CH2 | 1.42, m | 8 | 26.9, CH2 | 1.43, m | |
| 1.55, m | 1.69, m | |||||
| 12 | 40.7, CH2 | 1.40, m | 9, 18, 25 | 76.2, CH | 3.56, dd (11.8, 4.2) | |
| 1.78, d (9.8) | ||||||
| 13 | 37.8, C | 44.0, C | ||||
| 14 | 54.7, CH | 1.30, m | 15 | 24, 25 | 47.7, CH | 1.33, m |
| 15 | 24.2, CH2 | 2.07, m | 14, 16 | 16, 17 | 25.9, CH2 | 2.21, d (11.4) |
| 2.31, dd (20.4, 4.0) | 2.36, dt (20.2, 5.2) | |||||
| 16 | 136.6, CH | 6.84, d (3.3) | 15, 18 | 18, 20 | 143.1, CH | 7.28, m |
| 17 | 127.0, C | 125.8, C | ||||
| 18 | 57.9, CH | 2.47, m | 19 | 57.8, CH | 3.58, s | |
| 19 | 105.6, CH | 5.20, d (5.8) | 18 | 19-OMe | 204.0, CH | 9.86, d (3.6) |
| 20 | 171.0, C | 167.0, C | ||||
| 21 | 21.5, CH3 | 0.78, s | 22 | 21.4, CH3 | 0.77, s | |
| 22 | 33.5, CH3 | 0.84, s | 21 | 33.4, CH3 | 0.84, s | |
| 23 | 16.5, CH3 | 0.84, s | 17.0, CH3 | 0.88, s | ||
| 24 | 16.6, CH3 | 0.91, s | 16.8, CH3 | 0.89, s | ||
| 25 | 15.5, CH3 | 0.76, s | 16.0, CH3 | 0.90, s | ||
| 19-OMe | 57.9, CH3 | 3.56, s | ||||
| 20-OMe | 52.1, CH3 | 3.70, s | ||||




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download